Abstract
Tuberculosis remains a severe public health problem worldwide. Presently, genotyping is used for conducting epidemiologic and clinical studies on tuberculosis cases. We evaluated the efficacy of the repetitive sequence-based PCR (rep-PCR)-based DiversiLab™ system (bioMérieux, France) over the IS6110-restriction fragment length polymorphism analysis for detecting Mycobacterium tuberculosis. In all, 89 clinical M. tuberculosis isolates collected nationwide from Korea were used. The DiversiLab system allocated the 89 isolates to 8 groups with 1 unique isolate when a similarity level of 95% was applied. Seventy-six isolates of the Beijing family and 13 isolates of non-Beijing family strains were irregularly distributed regardless of rep-PCR groups. The DiversiLab system generated a rapid, sensitive, and standardized result. It can be used to conduct molecular epidemiologic studies to identify clinical M. tuberculosis isolates in Korea.
Tuberculosis (TB) remains a severe public health problem worldwide. In Korea, the prevalence of TB is still high despite multiple efforts to combat it [1, 2]. In addition, there has been a steady increase in the incidence of strains becoming resistant to drugs [3]. Genotyping is necessary for epidemiologic and clinical purposes and improves the understanding of the epidemiology and control of TB by providing information about transmission [4]. An epidemiologic tool, repetitive sequence-based PCR (rep-PCR), has been recognized as an effective method [5]. Rep-PCR is used to determine the similarity of bacterial strains at the genomic level as the repetitive sequences throughout the genome enable discrimination of interstrain variation on the basis of amplicon size and intensity. Rep-PCR greatly enhances assay reproducibility and strain discrimination compared with other PCR-based platforms. The commercial DiversiLab™ Microbial Typing System (bioMérieux, Marcy l'Etoile, France) [6] has the advantage of real-time data production and the convenience of partial automation [7]. IS6110-based restriction fragment length polymorphism (RFLP) is considered the gold standard and the most widely applied genotyping method for the molecular epidemiology of Mycobacterium tuberculosis [8].
In this study, we evaluated the DiversiLab rep-PCR-based system's discriminatory ability for M. tuberculosis by comparing it with previous data from IS6110-RFLP analysis [9]. We studied 89 clinical M. tuberculosis isolates collected from 11 university hospitals nationwide in Korea between 2008 and 2009 from epidemiologically unrelated patients. All 89 isolates had already been characterized by IS6110-RFLP [9]. According to the manufacturer's recommendations, the DiversiLab Fingerprinting Kit (Bacterial Barcodes, Inc., Huston, TX, USA) was used for rep-PCR amplification of non-coding intergenic repetitive elements in the genomic DNA of the isolates. Rep-PCR genomic typing was performed using the DiversiLab system. The DiversiLab system includes fragment separation using microfluidic chips (LabChip device; Caliper Technologies, Inc., Mountain View, CA, USA) and the Agilent B2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA, USA). Results were analyzed using DiversiLab software (version 3.3). Reports, including dendrograms, electropherograms, and virtual gel and scatterplot images, were generated automatically. To evaluate reproducibility, we performed the entire assay, including DNA extraction, PCR, and chip analysis on the bioanalyzer, in duplicate. The percent similarities of the dendrograms were analyzed by Pearson correlation. We used a single numerical index of discrimination on the basis of Simpson's index of diversity [10].
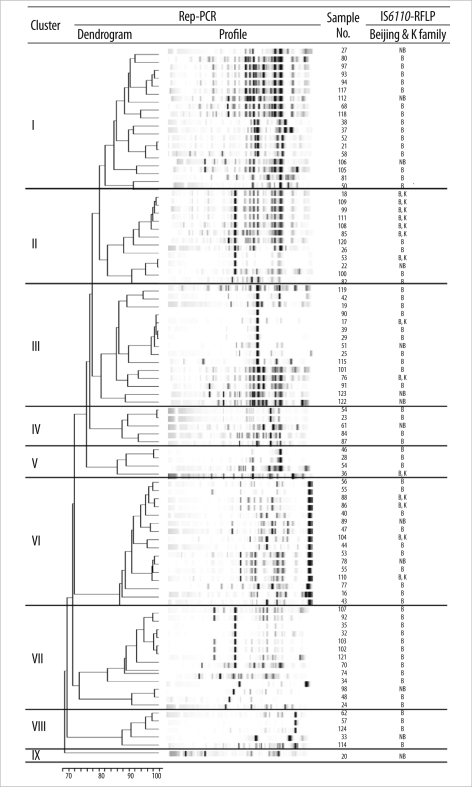
The results of the DiversiLab system defined 89 isolates in 8 groups and 1 unique isolate when a similarity level of 95% was applied (Fig. 1). One major group contained 18 isolates. Seventy-six isolates of the Beijing family and 13 isolates of non-Beijing family strains, previously defined by IS6110-RFLP [1], were distributed to each group. Therefore, this method was able to discriminate strain variations between Beijing family members. Among the 14 isolates of the K family, which showed unique RFLP patterns and are rather highly prevalent in Korea, 7 (50%) belonged to 1 major group. The reproducibility studies showed identical rep-PCR patterns for each duplicate test with the same strain. The results of cluster analysis did not correlate with any region of strain isolation.
Few publications are available on the performance of the DiversiLab system [11-13]. Cangelosi et al. [14] evaluated the system as an epidemiologic method for M. tuberculosis and M. avium complex, indicating that it can replace IS6110-RFLP for typing M. tuberculosis. In our current study, we showed the usefulness of the DiversiLab system by evaluating its efficacy with 89 M. tuberculosis isolates collected nationwide in Korea. The system displayed highly discriminatory results among Beijing family strains that have shown nearly identical IS6110-RFLP patterns. IS6110-RFLP is considered a gold standard method in strain typing for M. tuberculosis; however, discrimination of the Beijing family is unsatisfactory by using this method [15]. Therefore, IS6110-RFLP is less useful in Korea, where Beijing family strains are highly prevalent (87 to 95%) [9, 16]. The DiversiLab system can be a surrogate tool for epidemiologic analysis of M. tuberculosis in Korea. The ability to generate fingerprints in real time, together with the DNA amplification step, enables analysis in the early stages of the event; i.e., suspected cross contamination or outbreak.
Other molecular epidemiologic methods for M. tuberculosis strains have been evaluated. Mycobacterial interspersed repetitive units-variable number tandem repeats (MIRU-VNTR) is fairly rapid, requires only small amounts of DNA, and can easily be digitized to share data among laboratories [17]. Single-nucleotide polymorphism (SNP) analysis also provides a powerful strategy for large-scale molecular population studies examining phylogenetic relations among bacterial strains [18, 19]. However, the DiversiLab system provides results within 24 hr. Moreover, the DiversiLab system can generate a sensitive and standardized result and is reproducible and highly discriminatory. Thus, it can be used in the molecular epidemiologic investigation of clinical M. tuberculosis isolates in Korea.
Acknowledgement
All reagents used for DiversiLab analysis were purchased from, and technical support was freely provided by, bioMérieux Korea (Seoul, Korea).
References
1. Global tuberculosis control: key findings from the December 2009 WHO report. Wkly Epidemiol Rec. 2010; 85:69–80. PMID: 20210259.
2. Lönnroth K, Raviglione M. Global epidemiology of tuberculosis: prospects for control. Semin Respir Crit Care Med. 2008; 29:481–491. PMID: 18810682.

3. Espinal MA, Laszlo A, Simonsen L, Boulahbal F, Kim SJ, Reniero A, et al. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 2001; 344:1294–1303. PMID: 11320389.
4. Gopaul KK, Brown TJ, Gibson AL, Yates MD, Drobniewski FA. Progression toward an improved DNA amplification-based typing technique in the study of Mycobacterium tuberculosis epidemiology. J Clin Microbiol. 2006; 44:2492–2498. PMID: 16825370.
5. Dombek PE, Johnson LK, Zimmerley ST, Sadowsky MJ. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl Environ Microbiol. 2000; 66:2572–2577. PMID: 10831440.
6. Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. J Clin Microbiol. 2005; 43:199–207. PMID: 15634972.

7. Carretto E, Barbarini D, Farina C, Grosini A, Nicoletti P, Manso E. Use of the DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn Microbiol Infect Dis. 2008; 60:1–7. PMID: 17888611.
8. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993; 31:406–409. PMID: 8381814.
9. Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci. 2010; 25:1716–1721. PMID: 21165284.
10. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988; 26:2465–2466. PMID: 3069867.

11. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991; 19:6823–6831. PMID: 1762913.
12. Versalovic J, Kapur V, Mason EO Jr, Shah U, Koeuth T, Lupski JR, et al. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993; 167:850–856. PMID: 8450250.
13. Versalovic J, Kapur V, Koeuth T, Mazurek GH, Whittam TS, Musser JM, et al. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch Pathol Lab Med. 1995; 119:23–29. PMID: 7802548.
14. Cangelosi GA, Freeman RJ, Lewis KN, Livingston-Rosanoff D, Shah KS, Milan SJ, et al. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J Clin Microbiol. 2004; 42:2685–2693. PMID: 15184453.
15. van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995; 33:3234–3238. PMID: 8586708.
16. Shamputa IC, Lee J, Allix-Béguec C, Cho EJ, Lee JI, Rajan V, et al. Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J Clin Microbiol. 2010; 48:387–394. PMID: 20018816.
17. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006; 44:4498–4510. PMID: 17005759.
18. Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, et al. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot Minisequencing-based assay. J Clin Microbiol. 2010; 48:1758–1766. PMID: 20220173.
19. Choi GE, Jang MH, Cho HJ, Lee SM, Yi J, Lee EY, et al. Application of single-nucleotide polymorphism and mycobacterial interspersed repetitive units-variable number of tandem repeats analyses to clinical Mycobacterium tuberculosis isolates from Korea. Korean J Lab Med. 2011; 31:37–43. PMID: 21239869.
Fig. 1
Comparison of rep-PCR results with IS6110-RFLP patterns for 89 clinical Mycobacterium tuberculosis isolates. A scale for rep-PCR similarity (%) is shown at the bottom of the figure. IS6110-RFLP results are cited from reference 9.
Abbreviations: RFLP, restriction fragment length polymorphism; NB, strains of non-Beijing family; B, strains of Beijing family; K, strains of K family.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download