Abstract
The aminoglycoside 6'-N-acetyltransferases of type Ib (aac(6')-Ib) gene confers resistance to amikacin, tobramycin, kanamycin, and netilmicin but not gentamicin. However, some isolates harboring this gene show reduced susceptibility to amikacin. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommends a revision of the phenotypic description for isolates harboring the aac(6')-Ib gene. In this study, we determined the aminoglycoside susceptibility profiles of 58 AAC(6')-Ib-producing Enterobacter cloacae isolates. On the basis of the CLSI and EUCAST breakpoints, a large proportion (84.5% and 55.2%, respectively) of these 58 isolates were found to be susceptible to amikacin. However, among the isolates that were shown to be anikacin-susceptible according to the CLSI and EUCAST breakpoints, only 30.6% and 18.8% isolates, respectively, could be considered to have intermediate resistance on the basis of the EUCAST expert rules. Further studies should be conducted to determine the aminoglycoside susceptibility profiles of aac(6')-Ib-harboring isolates from various geographic regions and to monitor the therapeutic efficacy of amikacin in infections caused by these isolates.
Resistance to aminoglycosides is usually attributable to aminoglycoside-modifying enzymes. Among these, aminoglycoside 6'-N-acetyltransferases of type Ib [AAC(6')-Ib], is the most common cause of amikacin resistance among members of the family Enterobacteriaceae [1]. This enzyme can modify amikacin, tobramycin, kanamycin, and netilmicin but not gentamicin. Moreover, AAC(6')-Ib often coexists with other antibiotic-inactivating enzymes such as β-lactamases, carbapenemases, and other aminoglycosidases; therefore, clinical practitioners should be aware of its significance [2, 3].
The expert rules laid down by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) suggest that if an isolate of the family Enterobacteriaceae is intermediate or resistant to tobramycin and susceptible to gentamicin and amikacin, then its amikacin susceptibility status should be revised from "susceptible" to "intermediate" because production of acquired AAC(6')-I may not confer phenotypic amikacin resistance. In a previous study, we observed that over 40% of the Enterobacter cloacae isolates had the aac(6')-Ib gene [4]; however, many of the isolates with this gene were susceptible to amikacin [4]. Therefore, in this study, we determined the aminoglycoside susceptibility profiles of aac(6')-Ib-harboring E. cloacae isolates. Further, we investigated the aac(6')-Ib mutations (Leu119Ser, Leu120Ser, Glu167Ala, Phe171Ala, and Tyr166Ala) that are known to be associated with the loss of amikacin resistance [5-7].
We had previously collected 178 consecutive, non-duplicate isolates of E. cloacae from specimens obtained at 12 clinical microbiology laboratories in Korea between March 2005 and July 2005. The aac(6')-Ib gene was PCR amplified using 2 primers-5'-TTGCGATGCTCTATGGGCTA-'3 and 5'-CTCGAATGCCTGGCGTGTTT-'3-to obtain a 482-bp product [8]. In our previous study, all the 178 isolates were analyzed for the presence of aac(6')-Ib-cr [4]. Of the 74 E. cloacae isolates that were found to be positive for aac(6')-Ib, 58 were available for this study. For these 58 isolates, the minimum inhibitory concentrations (MICs) of amikacin (2-256 mg/L), kanamycin (2-256 mg/L), tobramycin (0.5-64 mg/L), and gentamicin (0.5-64 mg/L) were determined by the agar dilution method according to the CLSI guidelines [9].
To detect the mutations associated with the loss of amikacin resistance, the 482-bp PCR products were sequenced using a DNA Analyzer (Applied Biosystems, Foster City, CA, USA). To investigate the clonal relatedness of the isolates, pulsed-field gel electrophoresis (PFGE) was performed using a CHEF Mapper system (Bio-Rad Laboratories, Hercules, CA, USA). The whole-cell DNA was digested with XbaI, and the results were interpreted according to the criteria proposed by Tenover et al. [10].
Table 1 shows the antibiotic susceptibilities for the isolates. Of the 58 isolates harboring the aac(6')-Ib gene, 49 (84.5%) were susceptible to amikacin (≤16 mg/L); 2 (3.4%), to kanamycin (≤16 mg/L); 2 (3.4%), to tobramycin (≤4 mg/L); and 17 (29.3%), to gentamicin (≤4 mg/L) according to the CLSI breakpoints. According to the EUCAST breakpoints, 32 (52.2%) isolates were susceptible to amikacin (≤8 mg/L); 2 (3.4%), to tobramycin (≤2 mg/L); and 11 (19%), to gentamicin (≤2 mg/L). The distributions of the MICs of different aminoglycosides for the 58 E. cloacae isolates are shown in Fig. 1 and 2. Of the 49 amikacin-susceptible isolates (MIC ≤16 mg/L), only 2 isolates-KN7 and SO15-had the Leu119Ser mutation in the aac(6')-Ib gene. The KN7 isolate had 1 more amino acid change (Arg173Lys) in this gene. Findings of PFGE showed 10 different clones and no clonal relatedness among isolates from different clinical microbiology laboratories. However, clonal relatedness was observed among isolates from same hospitals: 2 out of 8, 2 out of 4, 2 out of 6, 3 and 2 out of 7, 2 out of 6, 2 and 2 out of 8, 4 out of 5, and 2 out of 2 isolates collected from 8 clinical microbiology laboratories. The isolates belonging to the same clone revealed identical PFGE pattern, but one isolate from hospital GM was related. We observed that 19 isolates belonging to 8 different clones were susceptible to amikacin (MIC ≤16 mg/L) and had different MICs for other aminoglycosides. However, 4 isolates belonging to the remaining 2 clones were resistant to all the tested aminoglycosides (MIC for amikacin and kanamycin, ≥256 mg/L; MIC for tobramycin and gentamicin, ≥64 mg/L) (data not shown).
A very high percentage of the aac(6')-Ib-harboring isolates were susceptible to amikacin (84.5% and 55.2% according to the CLSI and EUCAST breakpoints, respectively). However, among the isolates that were shown to be amikacin-susceptible according to the CLSI and EUCAST criteria, the phenotypes of only 30.6% (15/49) and 18.8% (6/32) isolates, respectively, could be revised according to the EUCAST expert rules because many of these amikacin-susceptible isolates were resistant to gentamicin. The gentamicin resistance may be because of gentamicin-modifying enzymes such as AAC(3)-I, AAC(3)-VI, AAC(2')-I, AAC(3)-IV, Ant (2')-I, and ACC(3)-II or impermeability. Since only 2 isolates had mutations associated with the loss of amikacin resistance, we think that the remaining isolates might have produced low-levels of AAC(6')-Ib. Low levels of AAC(6')-Ib could not confer resistance to amikacin and isepamicin in vitro but were able to efficiently modify the small amounts of drug entering the bacterial cell and thereby conferred resistance in vivo. Therefore, a previous study did not report any noticeable difference in the in vivo efficacies of amikacin or isepamicin for low- and high-level AAC(6')-Ib-producing organisms [11].
Our results show that the MICs of amikacin for many AAC(6')-Ib-producing isolates were below the susceptibility breakpoints recommended by the CLSI or EUCAST and that most of these susceptible isolates could not be considered to have intermediate amikacin resistance according to the above-mentioned EUCAST expert rules. Data on the epidemiology and optimal therapy of infections caused by these strains are scarce; therefore, further studies are needed to characterize these infections.
Acknowledgements
The authors wish to thank all the laboratories that provided isolates for this study. We are indebted to Kang Gyun Park for valuable technical assistance. This work was supported by a grant received from the Catholic Medical Center Research Foundation in 2008 and one received from St. Vincent's hospital in 2009.
References
1. Neonakis I, Gikas A, Scoulica E, Manios A, Georgiladakis A, Tselentis Y. Evolution of aminoglycoside resistance phenotypes of four gram-negative bacteria: an 8-year survey in a university hospital in Greece. Int J Antimicrob Agents. 2003; 22:526–531. PMID: 14602373.

2. Kim JY, Park YJ, Kwon HJ, Han K, Kang MW, Woo GJ. Occurrence and mechanisms of amikacin resistance and its association with β-lactamases in Pseudomonas aeruginosa: a Korean nationwide study. J Antimicrob Chemother. 2008; 62:479–483. PMID: 18606785.
3. Sabtcheva S, Kaku M, Saga T, Ishii Y, Kantardjjev T. High prevalence of the aac(6')-Ib-cr gene and its dissemination among Enterobacteriaceae isolates by CTX-M-15 plasmids in Bulgaria. Antimicrob Agents Chemother. 2009; 53:335–336. PMID: 19001110.
4. Kim SY, Park YJ, Yu JK, Kim YS, Han K. Prevalence and characteristics of aac(6')-Ib-cr in AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens: a multicenter study from Korea. Diagn Microbiol Infect Dis. 2009; 63:314–318. PMID: 19216942.
5. Casin I, Bordon F, Bertin P, Coutrot A, Podglajen I, Brasseur R, et al. Aminoglycoside 6'-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob Agents Chemother. 1998; 42:209–215. PMID: 9527761.
6. Rather PN, Munayyer H, Mann PA, Hare RS, Miller GH, Shaw KJ. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6'-N-acetyltransferase Ib and IIa proteins. J Bacteriol. 1992; 174:3196–3203. PMID: 1577689.
7. Shmara A, Weinsetel N, Dery KJ, Chavideh R, Tolmasky ME. Systematic analysis of a conserved region of the aminoglycoside 6'-N-acetyltransferase type Ib. Antimicrob Agents Chemother. 2001; 45:3287–3292. PMID: 11709299.
8. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006; 50:3953–3955. PMID: 16954321.
9. Clinical and Laboratory Standards Institute (CLSI). M100-S19. Performance standards for antimicrobial susceptibility testing; Nineteenth informational supplement. 2009. Wayne, PA: Clinical and Laboratory Standards Institute.
10. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–2239. PMID: 7494007.

11. Caulin E, Coutrot A, Carbon C, Collatz E. Resistance to amikacin and isepamicin in rabbits with experimental endocarditis of an aac(6')-Ib-bearing strain of Klebsiella pneumoniae susceptible in vitro. Antimicrob Agents Chemother. 1996; 40:2848–2853. PMID: 9124853.
Fig. 1
Distribution of the minimum inhibitory concentrations (MICs) of amikacin and kanamycin for the 58 Enterobacter cloacae isolates harboring the aac(6')-Ib gene*.
Black bars, amikacin; hatched bars, kanamycin.
*C1LSI-recommended MIC breakpoints for amikacin and kanamycin (S≤/I/R≥) are 16/32/64, and the corresponding EUCAST breakpoints for amikacin (S≤/R>) are 8/16.
Abbreviation: EUCAST; European Committee on Antimicrobial Susceptibility Testing.
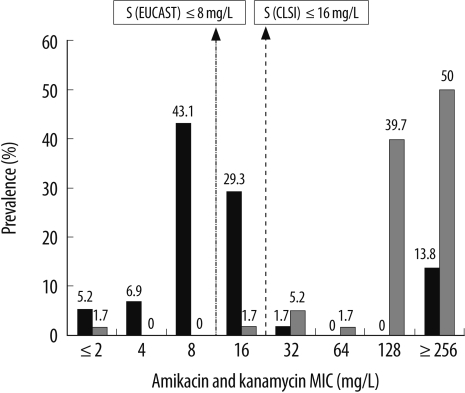
Fig. 2
Distribution of the minimum inhibitory concentrations (MICs) of tobramycin and gentamicin for the 58 Enterobacter cloacae isolates harboring the aac(6')-Ib gene*.
Black bars, tobramycin; hatched bars, gentamicin.
*CLSI-recommended MIC breakpoints for tobramycin and gentamycin (S≤/I/R≥) are 4/8/16, and the corresponding EUCAST breakpoints (S≤/R>) are 2/4.
Abbreviation: EUCAST; European Committee on Antimicrobial Susceptibility Testing.
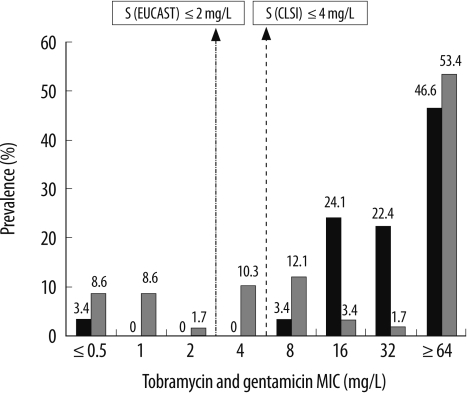
Table 1
Aminoglycoside susceptibilities of the 58 aac(6')-Ib-harboring Enterobacter cloacae isolates
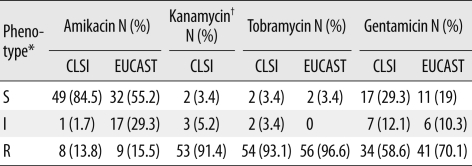
*CLSI-recommended minimum inhibitory concentration breakpoints for amikacin and kanamycin (S≤/I/R≥) are 16/32/64, and those for tobramycin and gentamicin are 4/8/16; the corresponding EUCAST breakpoint for amikacin (S≤/R>) are 8/16, and those for tobramycin and gentamicin are 2/4; †MIC breakpoint for kanamycin was not recommended by the EUCAST.
Abbreviations: S, susceptible; I, intermediate; R, resistant; EUCAST, European Committee on Antimicrobial Susceptibility Testing.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download