Abstract
Background
Members of the Acinetobacter calcoaceticus-baumannii (Acb) complex are important opportunistic bacterial pathogens and present significant therapeutic challenges in the treatment of nosocomial infections. In the present study, we investigated the integrons and various genes involved in resistance to carbapenems, aminoglycosides, and fluoroquinolones in 56 imipenem-nonsusceptible Acb complex isolates.
Methods
This study included 44 imipenem-nonsusceptible A. baumannii, 10 Acinetobacter genomic species 3, and 2 Acinetobacter genomic species 13TU strains isolated in Daejeon, Korea. The minimum inhibitory concentrations (MICs) were determined by Etest. PCR and DNA sequencing were used to identify the genes that potentially contribute to each resistance phenotype.
Results
All A. baumannii isolates harbored the blaOXA-51-like gene, and 21 isolates (47.7%) co-produced OXA-23. However, isolates of Acinetobacter genomic species 3 and 13TU only contained blaIMP-1 or blaVIM-2. Most Acb complex isolates (94.6%) harbored class 1 integrons, armA, and/or aminoglycoside-modifying enzymes (AMEs). Of particular note was the fact that armA and aph(3')-Ia were only detected in A. baumannii isolates, which were highly resistant to amikacin (MIC50≥256) and gentamicin (MIC50≥1,024). In all 44 A. baumannii isolates, resistance to fluoroquinolones was conferred by sense mutations in the gyrA and parC. However, sense mutations in parC were not found in Acinetobacter genomic species 3 or 13TU isolates.
Conclusions
Several differences in carbapenem, aminoglycoside, and fluoroquinolone resistance gene content were detected among Acb complex isolates. However, most Acb complex isolates (87.5%) possessed integrons, carbapenemases, AMEs, and mutations in gyrA. The co-occurrence of several resistance determinants may present a significant threat.
Acinetobacter spp. are ubiquitous in the environment and have emerged as important opportunistic pathogens that cause nosocomial infections [1]. Currently, more than 30 named and unnamed species of Acinetobacter have been identified by DNA-DNA hybridization [2]. Acinetobacter baumannii, which is the most frequently identified species from clinical infections and outbreaks, is genetically closely related and phenotypically similar to 2 other clinical species, namely, Acinetobacter genomic species (gen. sp.) 3 and 13TU and to Acinetobacter calcoaceticus, a soil bacterium. This close similarity led to the proposal that these species should be grouped together in the so-called A. calcoaceticus-A. baumannii (Acb) complex [3]. With the exception of A. calcoaceticus, all members of this complex have been reported to be involved in nosocomial infections and are known to have the ability to spread within hospitals [4].
Acb complex has now become a major cause of hospital-acquired infections worldwide due to its remarkable propensity to rapidly acquire determinants of resistance to a wide range of antibacterial agents [5]. In particular, increasing resistance to carbapenems has been observed worldwide in the past decade. Carbapenemase production is the best-described mechanism of resistance to carbapenems, and various metallo-β-lactamases (MBLs) and OXA-type carbapenemases have been isolated from Acb complex strains worldwide [6-8]. In some instances, carbapenemase genes, such as blaIMP, blaVIM, blaGIM, or blaSIM, are located within class 1 integrons and lead to carbapenem resistance. Multidrug-resistant Acb complex strains have been known to acquire their antimicrobial resistance genes via class 1 integrons that carry single or multiple gene cassettes [9].
Aminoglycoside resistance genes, such as acetyltransferase (aac), phosphotransferase (aph), and adenylyltransferase (aad), are also typically found within integron gene cassettes [10]. In addition, Acb complex strains carry various types of aminoglycoside-modifying enzymes (AMEs) and 16S rRNA methylases, which confer resistance to aminoglycosides [11].
Increases in the number of fluoroquinolone-resistant Acb complex strains are mediated primarily by spontaneous mutations in the quinolone resistance-determining regions (QRDRs) of either DNA gyrase (gyrA), topoisomerase IV (parC), or both [12].
Although the Acb complex has been recognized as an important opportunistic pathogen associated with life-threatening nosocomial infections and hospital outbreaks, there is a relative paucity of data on the number and type of resistance genes in Acb complex strains isolated in Daejeon, Korea. In the present study, we aimed to determine the genetic basis for multidrug resistance and the clonal relatedness of the Acb complex clinical isolates obtained from a university hospital in Daejeon. Molecular determinants enabling the multidrug resistant strains to exhibit co-resistance to carbapenems, aminoglycosides, and fluoroquinolones were investigated with a focus on integrons, carbapenemases, AMEs, 16S rRNA methylase, and mutations of the gyrA and parC genes.
A total of 56 consecutive and non-duplicate imipenem-nonsusceptible Acb complex isolates were collected from the university hospital laboratory in Daejeon, Korea, from January 2008 to December 2010. Acinetobacter spp. were identified by using the Vitek 2® automated ID system (BioMérieux, Marcy l'Etoile, France) and partial sequencing of the rpoB housekeeping gene, as described previously [13].
The MICs of Acb complex isolates for imipenem, meropenem, gentamicin, amikacin, ceftazidime, cefepime, and ciprofloxacin were determined using Etest (BioMérieux). Interpretation was based on the criteria approved by the CLSI [14]. Escherichia coli ATCC 25922 was used as the reference strain.
All Acb complex isolates were subjected to PCR and sequencing assays for the detection of antimicrobial resistance determinants and the identification of mutations associated with fluoroquinolone resistance. Specific primers for the detection of antimicrobial resistance determinants were used [15-18]. Chromosomal DNA was obtained from each target strain by using a genomic DNA purification kit (Promega, Madison, WI, USA) in accordance with standard protocols. PCR was performed using 50 ng of genomic DNA, 2.5 µL of 10× Taq buffer, 0.5 µL of 10 mM dNTP mix, 20 pmol of each primer, and 0.7 U of Taq DNA polymerase (SolGent, Daejeon, Korea), in a total volume of 25 µL. Each antimicrobial resistance determinant gene was amplified in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer Cetus Corp., Norwalk, CT, USA) by pre-denaturation of the reaction mixture at 95℃ for 5 min, followed by 35 cycles of 95℃ for 30 sec, 52℃ for 40 sec, and 72℃ for 30 sec, with a final extension at 72℃ for 5 min. The amplicons were purified with a PCR purification kit (SolGent) and were sequenced using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3730XL DNA analyzer (PE Applied Biosystems). Integron detection, characterization, and gene cassette amplification were carried out using previously described PCR methods [19].
In the REP-PCR method, the primer pair of REP1 (5'-IIIGCGCCGICATCAGGC-3') and REP2 (5'-ACGTCTTATCAGGCCTAC-3') was used to amplify putative REP-like elements in the genomic DNA [20]. Amplification reactions were performed in a final volume of 50 µL, containing 100 ng of chromosomal DNA, 5 µL of 10× Taq buffer, 1.0 µL of 10 mM dNTP mix, 1.5 U of Taq DNA polymerase (SolGent), and 50 pmol of each primer. The cycling conditions were as follows: an initial denaturation step at 95℃ for 5 min; followed by 30 cycles of 92℃ for 50 sec, 48℃ for 55 sec, and 70℃ for 5 min; and a final extension step at 70℃ for 10 min. The amplified products were separated via electrophoresis on 1.5% agarose gels containing ethidium bromide, and visualized using a BioDoc-14™ Imaging system (UVP, Cambridge, UK).
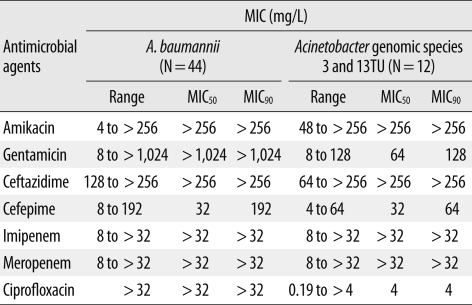
Isolates were assigned to 3 distinct Acinetobacter species on the basis of the results of rpoB gene analysis: A. baumannii (N=44), Acinetobacter gen. sp. 3 (N=10), and Acinetobacter gen. sp. 13TU (N=2). The distribution of MICs for imipenem, meropenem, amikacin, ceftazidime, and cefepime was similar among A. baumannii, Acinetobacter gen. sp. 3, and Acinetobacter gen. sp. 13TU. However, high-level resistance to gentamicin (MIC50≥1,024 mg/L) and ciprofloxacin (MIC50≥32 mg/L) was only seen in the A. baumannii isolates (Table 1).
Most of the Acb complex isolates (94.6%) contained class 1 integrons, which ranged between 1.8 and 3.0 kb in length. However, no class 2 or class 3 integrons were found within the cohorts. The gene cassettes found in this study were divided into 4 types by nucleotide sequencing (Fig. 1). The type 1 amplicon (3.0 kb), obtained in 10 Acinetobacter gen. sp. 3 isolates, carried aacA4-blaIMP-1-blaOXA-2 gene cassettes. The type 2 amplicon (3.0 kb), found in 2 Acinetobacter gen. sp. 13TU isolates, carried blaVIM-2-aacA7-aadA1 gene cassettes. The type 3 amplicon (1.8 kb), detected in only 1 Acinetobacter gen. sp. 13TU isolate, carried aac3-1-blaOXA-2-orfD gene cassettes. Forty-one A. baumannii isolates (93.2%) contained the type 4 amplicon (2.3 kb) carrying aacA4-catB8-aadA1 gene cassettes.
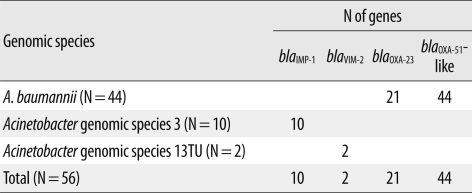
The distribution of carbapenem resistance genes among isolates of the 3 different Acinetobacter species is shown in Table 2. The blaOXA-23 gene was only amplified from 21 A. baumannii isolates (47.7%), all of which harbored the upstream insertion element ISAba1 and were highly resistant to imipenem (MICs≥32). The intrinsic β-lactamase gene, blaOXA-51-like, was amplified in all 44 A. baumannii isolates, and an upstream ISAba1 was identified in 39 strains (88.6%). Other OXA-type carbapenemase genes, such as blaOXA-24 and blaOXA-58-like, were not found in this study.
Two MBL genes, blaIMP-1 and blaVIM-2, were detected in Acinetobacter gen. sp. 3 and 13TU. The blaSIM gene was not detected in any of our isolates. All imipenem-nonsusceptible Acb complex isolates contained more than 1 carbapenemase gene.
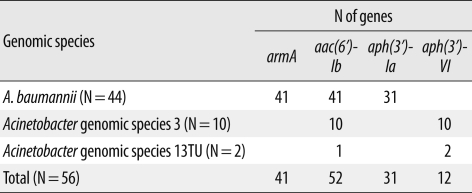
Three different genes encoding AMEs were identified: aac(6')-Ib was the most prevalent, found in 52 isolates, followed by aph(3')-Ia in 31 isolates, and aph(3')-VI in 12 isolates (Table 3). All 10 Acinetobacter gen. sp. 3 isolates contained aac(6')-Ib and aph(3')-VI, whereas 31 A. baumannii isolates had the combination of aac(6')-Ib/aph(3')-Ia. The 16S rRNA methylase gene armA was detected in 41 A. baumannii isolates, but rmtA and rmtB were not detected in any Acb complex isolates tested. The 41 A. baumannii isolates containing armA were highly resistant to amikacin (MIC50≥256 mg/L) and gentamicin (MIC50≥1,024 mg/L).
The 3 A. baumannii isolates that did not contain integrons or 16S rRNA methylase and AMEs had low-level aminoglycoside resistance, with an amikacin MIC50 of 4 mg/L and a gentamicin MIC50 of 8 mg/L.
All A. baumannii isolates had sense mutations at the 83rd residue (serine to leucine) in gyrA and at the 80th residue (serine to leucine or tryptophan) in parC, and had high-level ciprofloxacin resistance (MIC50≥32 mg/L). However, sense mutations in parC were not detected in Acinetobacter gen. sp. 3 and 13TU isolates. The sense mutations at the 83rd residue (serine to leucine) in gyrA were detected in only 8 Acinetobacter gen. sp. 3 isolates, which had low-level ciprofloxacin resistance (MIC50=4 mg/L). The 3 Acinetobacter gen. sp. isolates without sense mutations in either gyrA or parC were susceptible to ciprofloxacin (MIC50=0.19 mg/L).
To determine the clonality of all 56 Acb complex isolates, REP-PCR was performed on genomic DNA. The 44 A. baumannii isolates displayed only 2 REP-PCR types, designated type A and type B (Fig. 2). However, the Acinetobacter gen. sp. 3 and 13TU isolates showed diverse band patterns.
This study analyzed various genes of Acb complex isolates that are responsible for resistance to carbapenems, aminoglycosides, and fluoroquinolones. Previous studies focused mainly on carbapenem or aminoglycoside resistance [11]. We found differences in the genetic characteristics of isolates from 3 different Acinetobacter species in terms of their carbapenem, aminoglycoside, and fluoroquinolone resistance.
Our study identified several differences in carbapenem resistance genes among Acinetobacter spp. isolates. OXA-type carbapenemase genes (blaOXA-23 and blaOXA-51-like) were only observed in A. baumannii isolates, while MBL genes (blaIMP-1 or blaVIM-2) were only present in Acinetobacter gen. sp. 3 or 13TU isolates. This distribution is in accordance with previous reports of resistance genes among Acinetobacter spp. [21, 22]. The presence of ISAba1 upstream of blaOXA genes provides a promoter sequence enhancing their expression [23]. In isolates possessing blaOXA-23, ISAba1 was consistently associated with blaOXA-23 and not with the blaOXA-51-like gene, suggesting that coexpression of these 2 genes does not occur. The dissemination of A. baumannii isolates harboring the blaOXA-23 gene has been previously reported in Korea [15, 24]. In addition, class 1 integrons harboring MBL genes were detected in Acinetobacter gen. sp. 3 and 13TU isolates. A 3.0-kb class 1 integron containing blaIMP-1 with aacA4 and blaOXA-2 was described in A. baumannii (EF375699) and A. junii (EU014166) isolates recovered from Korea in 2007. In our previous study, we reported that multidrug resistant A. baumannii isolates contained class 1 integrons harboring blaIMP-1 with aacA4 and blaOXA-2 [15]. However, an A. baumannii isolate harboring blaIMP-1 was not detected in the present study. A 3.0-kb class 1 integron containing blaVIM-2 with aacA7 and aadA1 was described in A. baumannii (AF324464) recovered from Korea in 2002 [25].
In our study, significant variation was seen in the distribution of aminoglycoside resistance genes among Acinetobacter gen. sp. isolates. The armA and aph(3')-Ia genes were only detected in A. baumannii isolates, whereas aph(3')-VI was only detected in Acinetobacter gen. sp. 3 and 13TU isolates. In particular, A. baumannii isolates containing armA showed high-level gentamicin resistance (MIC50≥1,024 mg/L). Unlike AMEs, which vary in their substrate ranges, 16S rRNA methylases confer high-level resistance to almost all clinically important aminoglycosides [11]. The armA gene was previously detected in A. baumannii, P. aeruginosa, E. coli, K. pneumoniae, and S. marcescens [26], but has not been detected in other Acinetobacter gen. sp. so far.
A major mechanism of fluoroquinolone resistance in gram-negative bacteria involves changes in the structure of the drug targets, DNA gyrase and DNA topoisomerase IV [27]. In particular, amino acid substitutions in both the GyrA and ParC polypeptides are consistent with a high-level fluoroquinolone resistant phenotype [28]. In this study, all 44 A. baumannii isolates harboring sense mutations in both gyrA and parC showed high-level resistance to ciprofloxacin (MIC50≥32 mg/L). These results are similar to those reported in our study of multidrug resistant A. baumannii isolates [15].
In addition, identical banding patterns on REP-PCR profiles were only seen among the A. baumannii isolates. Our results suggest that there is both clonal and horizontal spread of resistance genes between imipenem-nonsusceptible A. baumannii isolates in the university hospital in Daejeon.
Despite the increasingly frequent discovery of multidrug resistant Acinetobacter spp. isolates in Korea, there is a relative paucity of information regarding the antimicrobial resistance of this gram-negative bacillus in Daejeon. Although we found differences in genetic characteristics of isolates from 3 different Acinetobacter species, most (87.5%) of them possessed integrons, AMEs, carbapenemases, and mutations in gyrA. The co-occurrence of several resistance determinants may present a significant threat.
References
1. Bergogne-Bérézin E. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996; 9:148–165. PMID: 8964033.
2. La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006; 44:827–832. PMID: 16517861.
3. Gerner-Smidt P, Tjernberg I. Acinetobacter in Denmark: II. Molecular studies of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. APMIS. 1993; 101:826–832. PMID: 8286091.
4. van den Broek PJ, van der Reijden TJ, van Strijen E, Helmig-Schurter AV, Bernards AT, Dijkshoorn L. Endemic and epidemic Acinetobacter species in a university hospital: an 8-year survey. J Clin Microbiol. 2009; 47:3593–3599. PMID: 19794057.
5. Gootz TD, Marra A. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev Anti Infect Ther. 2008; 6:309–325. PMID: 18588496.
6. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii mechanisms and epidemiology. Clin Microbiol Infect. 2006; 12:826–836. PMID: 16882287.
7. Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007; 2:501–512. PMID: 17927473.

8. Mugnier PD, Poirel L, Nordmann P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol. 2009; 191:2414–2418. PMID: 19136598.
9. Petersen A, Guardabassi L, Dalsgaard A, Olsen JE. Class I integrons containing a dhfrI trimethoprim resistance gene cassette in aquatic Acinetobacter spp. FEMS Microbiol Lett. 2000; 182:73–76. PMID: 10612734.
10. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004; 53:1233–1240. PMID: 15585503.
11. Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–190. PMID: 19361944.
12. Vila J, Ruiz J, Goñi P, Marcos A, Jimenez de Anta T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995; 39:1201–1203. PMID: 7625818.
13. Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology. 2009; 155:2333–2341. PMID: 19389786.
14. Clinical and Laboratory Standards Institute. M100-S20. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. 2010. Wayne, PA: Clinical and Laboratory Standards Institute.
15. Koo SH, Kwon KC, Cho HH, Sung JY. Genetic basis of multidrug-resistant Acinetobacter baumannii clinical isolates from three university hospitals in Chungcheong province, Korea. Korean J Lab Med. 2010; 30:498–506. PMID: 20890082.
16. Akers KS, Chaney C, Barsoumian A, Beckius M, Zera W, Yu X, et al. Aminoglycoside resistance and susceptibility testing errors in Acinetobacter baumannii-calcoaceticus complex. J Clin Microbiol. 2010; 48:1132–1138. PMID: 20107089.
17. Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, De Mendonca R, et al. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother. 2007; 59:459–464. PMID: 17224412.
18. Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007; 45:544–547. PMID: 17093019.

19. Shin SY, Kwon KC, Park JW, Song JH, Ko YH, Sung JY, et al. Characteristics of aac(6')-Ib-cr gene in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from Chungnam area. Korean J Lab Med. 2009; 29:541–550. PMID: 20046086.
20. Bou G, Cerveró G, Domínguez MA, Quereda C, Martínez-Beltrán J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000; 6:635–643. PMID: 11284921.
21. Lee JH, Choi CH, Kang HY, Lee JY, Kim J, Lee YC, et al. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J Antimicrob Chemother. 2007; 59:633–639. PMID: 17339277.
22. Lee K, Kim MN, Choi TY, Cho SE, Lee S, Whang DH, et al. Wide dissemination of OXA-type carbapenemases in clinical Acinetobacter spp. isolates from South Korea. Int J Antimicrob Agents. 2009; 33:520–524. PMID: 19091520.
23. Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006; 258:72–77. PMID: 16630258.
24. Sung JY, Kwon KC, Park JW, Kim YS, Kim JM, Shin KS, et al. Dissemination of IMP-1 and OXA type β-lactamase in carbapenem-resistant Acinetobacter baumannii. Korean J Lab Med. 2008; 28:16–23. PMID: 18309251.
25. Yum JH, Yi K, Lee H, Yong D, Lee K, Kim JM, et al. Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla(VIM-2) gene cassettes. J Antimicrob Chemother. 2002; 49:837–840. PMID: 12003980.
26. Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, et al. Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010; 29:1349–1353. PMID: 20614151.

27. Akasaka T, Tanaka M, Yamaguchi A, Sato K. Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob Agents Chemother. 2001; 45:2263–2268. PMID: 11451683.
28. Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008; 46:2499–2507. PMID: 18524965.
Fig. 1
Schematic representation of gene cassette structures located in the class 1 integron isolated from Acinetobacter calcoaceticus-baumannii complex isolates. The horizontal arrows indicate the translation orientation of the genes.
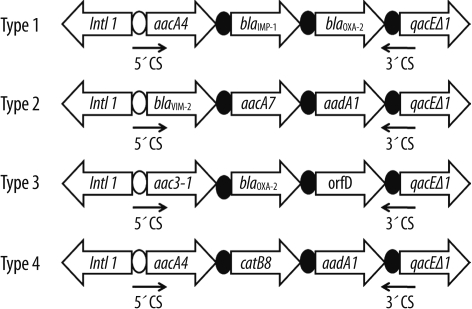
Fig. 2
Repetitive extragenic palindromic (REP)-PCR patterns of genomic DNA from imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolates. Lane M, 1-kb DNA size marker; Ab, Acinetobacter baumannii; Ag, Acinetobacter genomic species 3.
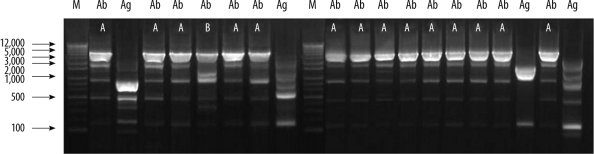
Table 1
The MIC distribution of 7 antimicrobial agents for the 56 A. calcoaceticus-baumannii complex isolates as determined by Etest




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download