Abstract
Background
Human bocavirus (HBoV) is a newly identified viral pathogen, and its clinical epidemiology and significance in respiratory infections have not yet been completely elucidated. We investigated the prevalence of HBoV infection and the association between viral (HBoV) load and clinical features of the infection in patients of all age-groups.
Methods
Nasopharyngeal aspirates from patients with symptoms of respiratory infection were tested for presence of HBoV by using real-time polymerase chain reaction. HBoV-positive patients were categorized into low- and high-viral-load groups using 1.0×106 copies/mL as the threshold value of viral load.
Results
Detection rate of HBoV was 4.8% (N=93) in a total of 1,926 samples with peak incidence of infection being observed in patients aged 6-12 months. HBoV infection was more frequently observed in young children, especially, in children aged less than 5 yr, and the HBoV load decreased with increase in age. HBoV was codetected with other respiratory viruses in 17 (18.3%) of the 93 HBoV-positive patients and 15 patients (88.2%) belonged to the low-viral-load group. Patients infected with HBoV alone showed a higher viral load than those patients in whom HBoV was codetected with other respiratory viruses (median load, 3.78×105 copies/mL vs. 1.94×104 copies/mL, P=0.014). Higher pulse rate (P=0.007) and respiratory rate (P=0.021) were observed in patients with a high-viral-load.
Human bocavirus (HBoV) was newly discovered by Allander et al. [1] and was stated to be the possible causative agent of respiratory illness. Phylogenetic analysis of the complete genome of HBoV revealed that the virus is most closely related to canine minute virus and bovine parvovirus, which are members of the Bocavirus genus of the Parvoviridae family [1].
HBoV was found in 1.5-11.3% of respiratory samples investigated worldwide [2-5]. HBoV may be a causative agent of respiratory tract infections. However, some investigators argue that the association between HBoV and respiratory tract diseases remains unproven because of the high rate of codetection of HBoV with other respiratory pathogens [5]. Its clinical epidemiology and role in respiratory infection have not yet been fully elucidated.
Although recently it was found that HBoV could be cultured in differentiated human airway epithelial cells [6], routine viral culturing of HBoV remains difficult. Real-time PCR has been used to estimate viral load and its usefulness has been proved as an indicator of the degree of active viral infection, interactions between the virus and the host, and the role of viral reactivation or persistence in the progression of disease [7]. In this study, we investigated the epidemiology of HBoV and the clinical features of respiratory infection associated with HBoV, especially in terms of HBoV load.
From November 2005 to October 2006, 1,926 specimens from 1,815 patients were randomly selected among the 5,891 samples collected from patients referred to the Korea University Guro Hospital for screening for respiratory viruses. Among the patients, 59.0% were men, and the ages of the patients ranged from 1 day to 86.0 yr (mean=6.1 yr, median=26 months); children aged 5 yr or less constituted 75.3% of the study population. The nasopharyngeal aspirates (NPAs) obtained from all patients were stored in a viral transport medium and quickly delivered to the laboratory by using wet ice. R-Mix culture system (Diagnostic HYBRIDS, Athens, OH, USA) was used to screen the samples for presence of 5 respiratory viruses (influenza A and B viruses, parainfluenza viruses, respiratory syncytial virus, and adenovirus).
Total nucleic acid was extracted from 100 µL of the sample by using Instagene Matrix Kit (Bio-Rad, Hercules, CA, USA). PCR primers were designed using Beacon Designer software (Premier Biosoft International, Palo Alto, CA, USA) in the conserved region of the NS-1 coding region of the HBoV genome by using the HBoV ST2 sequence (Genbank accession number: DQ000496). The forward primer (5'-GCAAATCTCTTCTGGCTACACG-3') and the reverse primer (5'-CCTCTGCGATCTCTATATTGAAGG-3') were targeted at a portion of the HBoV NS-1 gene. The conventional PCR reaction mixture consisted of 0.2 pg/µL forward and reverse primers, 2.5 mM dNTPs, 50 mM KCl, 1.5 mM MgCl2, 5 U of Taq polymerase, and 3 µL of extracted DNA in a final volume of 25 µL. The PCR cycling conditions consisted of 35 cycles (involving reaction at 30 sec at 94℃, 30 sec at 60℃, and 30 sec at 72℃) after the preheating step of 3 min at 94℃. All the PCR products obtained from positive reactions were sequenced completely to confirm sequence specificity.
To obtain standard curves for absolute quantification, the PCR product of HBoV was cloned into a plasmid vector pGEM®-T Easy Vector (Promega, Madison, WI, USA), purified using a QIAprep mini prep kit (Qiagen Inc., Valencia, CA, USA), and quantified using UV spectroscopy (1 genomic copy=5×10-12 µg). Serial 10-fold dilutions of the clon-ed plasmid were prepared to generate the standard curves.
Real-time PCR for viral load was performed on the specimens positive for HBoV by conventional PCR. A TaqMan probe (5'-ATGTTGCCGCCAGTAACTCCACCC-3') was labeled at the 5' ends with the reporter molecule FAM and at the 3' ends with Black Hole Quencher 1 (Biosearch Technologies, Inc., Novato, CA, USA). The assay was performed using Rotor-Gene 6000 (Corbett Life Science, Sydney, Australia) and the standard protocol of TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA, USA); each 25 µL sample of the reaction mixture contained 10 pg/µL of the forward and the reverse primers and 3 µL of the extracted DNA. Amplification conditions consisted of reactions for 3 min at 50℃, 3 min at 94℃, and 40 cycles of 30 sec at 94℃, 30 sec at 60℃ and 30 sec at 72℃ maintained for 3 min. Detection limit of real-time PCR for HBoV was 1.3×103 copies/mL, which corresponded to 33 copies per reaction.
Statistical analysis was performed using the SPSS software (version 10, SPSS Inc., Chicago, IL, USA), and graphs were prepared using Prism software (version 4.0, GraPad Software, Inc., San Diego, CA, USA). Mann-Whitney U test, Chi-squared or Fisher's exact test were performed to assess the significance. A P value <0.05 was considered statistically significant for all the tests.
A total of 1,926 samples of patients with respiratory sym-ptoms were included in the present study. Ninety-three (4.8%) samples were found to be positive for HBoV by PCR and subsequent sequencing. Other respiratory viruses detected during the study period were as follows: influenza A virus (IFA), 56 patients (2.9%); influenza B virus (IFB), 50 patients (2.6%); parainfluenza viruses (PIV), 142 patients (7.4%); respiratory syncytial virus (RSV), 97 patients (5.0%); and adenovirus (ADV), 40 patients (2.1%). HBoV was more prevalent in men (73.1%) than in women (P=0.005).
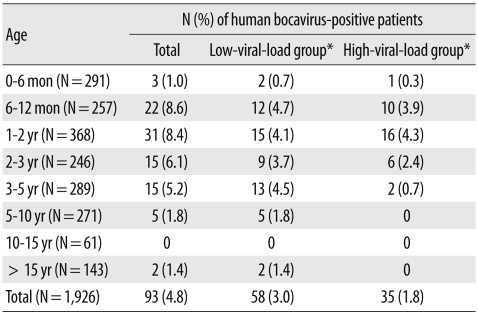
HBoV was detected in patients ranging from 3 months to 65.6 yr (mean=36.2 months, median=19 months) with a peak prevalence between the ages of 6 and 12 months (8.6%, 22/257). Children aged 5 yr or less constituted 92.5% (86/93) of the HBoV-positive patients (Table 1).
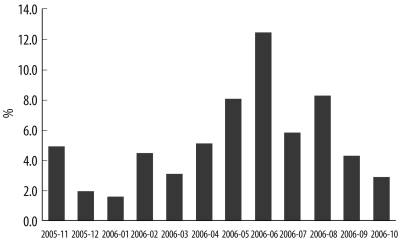
HBoV was detected in the samples obtained throughout the course of the study with the detection rate being the highest in June (16/129, 12.4%), followed by August (9/110, 8.2%) and May (13/162, 8.0%). From April to June 2006, 43% of the HBoV-positive cases were observed (Fig. 1).
All the 93 HBoV-positive cases detected by conventional PCR were also confirmed by real-time PCR performed using the TaqMan probe. The viral load detected in the HBoV-positive samples by using real-time PCR was in the range of 1.3×103-4.6×109 copies/mL (median, 1.82×105 copies/mL). The HBoV-positive cases were categorized in 2 groups: low-viral-load group (viral load ≤1.0×106 copies/mL, N=58) and high-viral-load group (viral load >1.0×106 copies/mL, N=35) (Table 1). The HBoV-positive patients aged less than 3 yr had a significantly higher viral load than that in the patients aged more than 3 yr (P=0.001).
The other respiratory viruses found in 17 (18.3%) of the total HBoV-positive samples were as follows: IFA, 1 sample; IFB, 1 sample; PIV, 10 samples; RSV, 4 samples; and ADV, 1 sample. Most of the cases (88.2%, 15/17) belonged to the low-viral-load group. RSV was isolated from the remaining 2 samples with HBoV copy numbers of 1.09×106 copies/mL and 2.65×108 copies/mL.
Patients positive for HBoV alone had a higher viral load than that in the patients who were positive for both HBoV and another respiratory virus (median 3.78×105 copies/mL vs. 1.94×104 copies/mL, P=0.014). A high-viral-load was almost exclusively seen in the HBoV-positive patients alone (94.3%, 33/35) (Fig. 2).
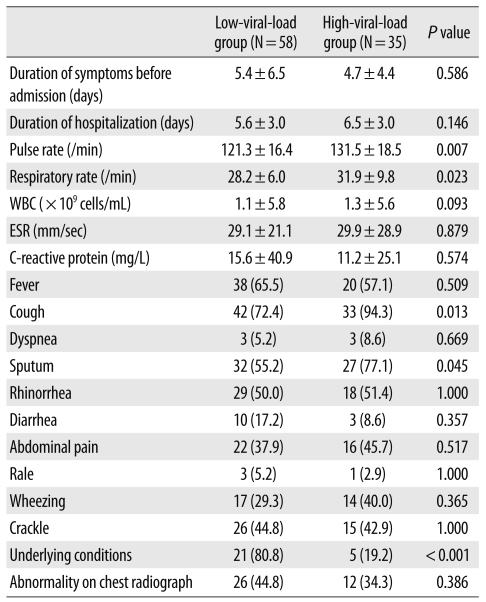
The high-viral-load group had a significantly higher pulse rate and respiratory rate than the corresponding rates in the low-viral-load group (P=0.007 and P=0.0231, respectively; Table 2). Although the duration of hospital stay was not significantly different between the 2 groups (Table 2), in cases of patients with less than 10 days of hospital stay, the high-viral-load group had a longer hospital stay than the low-viral-load group did (5.2±1.5 days vs. 4.1±1.4 days, P=0.009). Most of the other clinical characteristics had no significant correlation with the viral load of HBoV. The HBoV-positive patients presented with cough (80.6%), sputum (63.4%), fever (62.4%), rhinorrhea (50.5%), crackle (44.1%), wheezing (33.3%), diarrhea (14.0%), and dyspnea (6.5%). No significant difference was observed in the HBoV viral loads in the cases of upper and lower respiratory tract infections (P=0.077). Twenty-one out of 26 patients with underlying conditions had low-viral-loads.
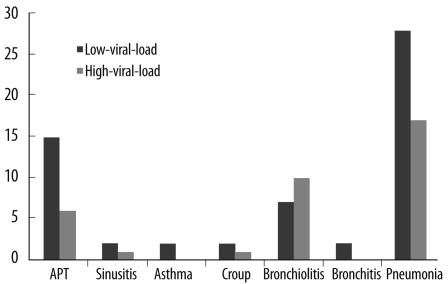
Clinical examination of the HBoV-positive patients showed pneumonia, bronchiolitis, bronchitis, croup, asthma, sinusitis, and pharyngotonsilitis (Fig. 3). HBoV was detected in 8.4% (N=48) and 5.6% (N=45) of the samples obtained from patients with and without pneumonia, respectively (N=572 and 805, respectively; P=0.049). However, no significant difference was observed in the viral load between the patients with and without pneumonia (P>0.05).
Ninety-three (4.8%) of the 1,926 nasopharyngeal aspirates obtained from patients of all age-groups were positive for HBoV. Our detection rate is similar to that stated in other reports [2, 3, 8]. Generally, HBoV is detected in fewer than 8% of respiratory specimens [1, 8-13]; however, higher detection rates ranging from 10.3% to 19% have been reported [5, 14, 15].
To investigate the epidemiological association of respiratory infection with viral load, HBoV-positive patients were categorized into low- and high-viral-load groups by using 1.0×106 copies/mL as a threshold value. The detection rate of HBoV infection was at its peak in the first year of life (rate of detection, 8.6% between the ages of 6 and 12 months), as in the cases of RSV or PIV infection. Most of the HBoV-positive patients aged less than 3 yr belonged to the high-viral-load group. The detection rate was lower (1.5%) in patients aged more than 10 yr, and these patients belonged exclusively to the low-viral-load group. HBoV is rarely detected in adults except in cases of immunosuppression [10, 16, 17]. The lower detection rate and viral load of HBoV in older patients may be attributed to immunity acquired from an infection at a younger age. A seroepidemiologic study of HBoV showed that 5.6-83.3% of children aged 6 months-3 yr were seropositive for HBoV [18]. Lau et al. [19] suggested that HBoV infection might develop only once because of the subsequent development of life-long immunity conferred by neutralizing antibodies produced in response to the infection.
The frequency of HBoV codetection with other respiratory viruses was 18.3% in the HBoV-positive samples and was lower than the previously published data [4, 20]. This difference in the codetection frequency is attributed to the different detection methods; molecular diagnostic methods were used for the detection of other respiratory viruses in the other studies whereas we used virus culturing. Among the 17 HBoV-positive patients who were also positive for infection with other viruses, 10 showed PIV infection. The high association of HBoV with PIV seems to be attributed to the high prevalence of PIV infection in 2006 (22.8% in May). Interestingly, almost all cases (except two) positive for both HBoV and another respiratory virus belonged to the low-viral-load group. As the virus culture was used for the detection of major respiratory viruses, the isolated virus could be the main causative agent of respiratory illness. Therefore, the presence of low copy number of HBoV, detected by molecular method, may indicate prolonged viral shedding or an asymptomatic infection. Recently, prolonged presence of HBoV in NPAs has been reported [21]. These results suggest that single HBoV infection in the high-viral-load group may play an active role in respiratory infection. These findings are consistent with a Norwegian study that reported detection of HBoV alone and a high-viral-load were associated with respiratory tract infection [20]. In that study, patients with a high-viral-load in NPAs developed viremia more frequently than the patients with a moderate-or a low-viral-load did. In our study, HBoV-positive patients in the high-viral-load group showed significantly higher pulse rates and respiratory rates than the in the low-viral-load group. These findings also support the idea that a high-viral-load may be associated with a respiratory infection.
Previous studies have reported that HBoV infection was more prevalent among individuals who had other respiratory viruses [10, 22]. In a study performed in Hong Kong [19], a higher detection rate of HBoV was observed in NPAs positive for common respiratory viruses than in those that were negative for the same. However, in our study, similar detection rate of HBoV was observed in the samples positive and negative for other respiratory viruses in the R-mix culture (Data are not shown).
Previous studies showed that cases of HBoV infection were found throughout the year with a peak incidence rate in the winter season [10, 13, 23]. However, in our study, cases of HBoV infection were detected most frequently during the spring season. This finding is similar to those of reports from Korea [4, 24]. This seasonal difference in the incidence of HBoV infection may be attributed to regional and temporal differences.
Bastien et al. [8] suggested that risk factors for severe HBoV infection appear to be similar to those for RSV infection (prematurity, congenital heart disease, and asthma). Thirty percent (26/93) of the HBoV-positive patients had underlying conditions such as heart disease, asthma, allergy, preterm birth, and a history of convulsions; most of these patients (80.8%) showed a low-viral-load. Persistent HBoV shedding for more than 1 month is observed in both respiratory and fecal specimens obtained from patients with significant underlying diseases [19]. Although these finding were not fully understood, it is postulated to be a result of underlying immunosuppression [19].
In summary, HBoV infection was more prevalent in young children. Patients positive for HBoV alone mainly constituted the high-viral-load group. Most of the HBoV-positive patients with infection caused by other respiratory viruses belonged to the low-viral-load group. These findings suggest that HBoV may be associated with a respiratory infection.
References
1. Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular scr-eening of respiratory tract samples. Proc Natl Acad Sci USA. 2005; 102:12891–12896. PMID: 16118271.
2. Ma X, Endo R, Ishiguro N, Ebihara T, Ishiko H, Ariga T, et al. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006; 44:1132–1134. PMID: 16517912.

3. Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mack-ay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006; 35:99–102. PMID: 16257260.

4. Choi EH, Lee HJ, Kim SJ, Eun BW, Kim NH, Lee JA, et al. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin Infect Dis. 2006; 43:585–592. PMID: 16886150.

5. Weissbrich B, Neske F, Schubert J, Tollmann F, Blath K, Blessing K, et al. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect Dis. 2006; 6:109. PMID: 16834781.

6. Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol. 2009; 83:7739–7748. PMID: 19474096.

7. Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004; 10:190–212. PMID: 15008940.

8. Bastien N, Chui N, Robinson JL, Lee BE, Dust K, Hart L, et al. Detection of human bocavirus in Canadian children in a 1-year study. J Clin Microbiol. 2007; 45:610–613. PMID: 17122013.

9. Kahn J. Human bocavirus: clinical significance and implications. Curr Opin Pediatr. 2008; 20:62–66. PMID: 18197041.

10. Manning A, Russell V, Eastick K, Leadbetter GH, Hallam N, Templeton K, et al. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J Infect Dis. 2006; 194:1283–1290. PMID: 17041855.

11. Kesebir D, Vazquez M, Weibel C, Shapiro ED, Ferguson D, Landry ML, et al. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006; 194:1276–1282. PMID: 17041854.

12. Naghipour M, Cuevas LE, Bakhshinejad T, Dove W, Hart CA. Human bocavirus in Iranian children with acute respiratory infections. J Med Virol. 2007; 79:539–543. PMID: 17385723.

13. Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006; 78:1232–1240. PMID: 16847968.

14. Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, et al. Human bocavirus and acute wheezing in children. Clin Infect Dis. 2007; 44:904–910. PMID: 17342639.

15. Kleines M, Scheithauer S, Rackowitz A, Ritter K, Häusler M. High prevalence of human bocavirus detected in young children with severe acute lower respiratory tract disease by use of a standard PCR protocol and a novel real-time PCR protocol. J Clin Microbiol. 2007; 45:1032–1034. PMID: 17215343.

16. Bastien N, Brandt K, Dust K, Ward D, Li Y. Human bocavirus infection, Canada. Emerg Infect Dis. 2006; 12:848–850. PMID: 16704852.

17. Fry AM, Lu X, Chittaganpitch M, Peret T, Fischer J, Dowell SF, et al. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis. 2007; 195:1038–1045. PMID: 17330795.

18. Endo R, Ishiguro N, Kikuta H, Teramoto S, Shirkoohi R, Ma X, et al. Seroepidemiology of human bocavirus in Hokkaido prefecture, Japan. J Clin Microbiol. 2007; 45:3218–3223. PMID: 17699639.

19. Lau SK, Yip CC, Que TL, Lee RA, Au-Yeung RK, Zhou B, et al. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J Infect Dis. 2007; 196:986–993. PMID: 17763318.

20. Christensen A, Nordbø SA, Krokstad S, Rognlien AG, Døllner H. Human bocavirus in children: mono-detection, high-viral-load and viraemia are associated with respiratory tract infection. J Clin Virol. 2010; 49:158–162. PMID: 20833582.
21. Blessing K, Neske F, Herre U, Kreth HW, Weissbrich B. Prolonged detection of human bocavirus DNA in nasopharyngeal aspirates of children with respiratory tract disease. Pediatr Infect Dis J. 2009; 28:1018–1019. PMID: 19730155.

22. Franz A, Adams O, Willems R, Bonzel L, Neuhausen N, Schweizer-Krantz S, et al. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol. 2010; 48:239–245. PMID: 20646956.

23. Arnold JC, Singh KK, Spector SA, Sawyer MH. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin Infect Dis. 2006; 43:283–288. PMID: 16804840.

24. Chung JY, Han TH, Kim SW, Kim CK, Hwang ES. Detection of viruses identified recently in children with acute wheezing. J Med Virol. 2007; 79:1238–1243. PMID: 17597481.

Fig. 2
HBoV viral load distribution of cases with HBoV infection alone and of cases in which HBoV was codetected with other viruses. The dotted line indicates the cut-off between the high-and the low-viral-load groups. The dashed lines indicate the median of each group.
Abbreviation: HBoV, human bocavirus.
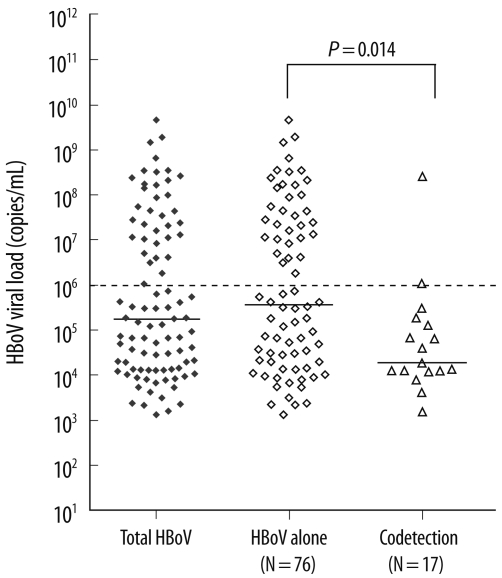
Fig. 3
Distribution of viral load in HBoV-positive patients according to diseases.
Abbreviation: HBoV, human bocavirus.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download