Abstract
Determination of the copy number of the survival motor neuron (SMN) gene is important for detecting spinal muscular atrophy (SMA) carriers and compound heterozygous patients. Multiplex ligation-dependent probe amplification (MLPA) assay is a simple and efficient technique used for detecting variations in the copy numbers of different genes. Race- and ethnicity-based variation in the SMA carrier frequency and the ‘2+0’ genotype of SMN1 are important factors that should be considered when estimating the risk of being an SMA carrier. Since SMN2 plays a disease-modifying role, accurate determination of SMN2 copy numbers in SMA patients can serve as a useful prognostic tool. Therefore, information on the SMN2 genotype distributions in normal populations will be helpful in selecting appropriate reference samples for MLPA analysis. To determine SMA carrier frequencies and SMN genotype distribution, we determined the copy numbers of SMN1 and SMN2 genes using the MLPA assay in 100 unrelated Korean individuals with no family history of SMA. The frequency of SMA carriers in the Korean population appears to be 1 in 50, which indicates that the prevalence of SMA among Koreans is the same as that among individuals in the Western countries. Two of the 100 normal individuals enrolled in this study showed 3 copies of the SMN1 gene. Therefore, 1.0% of the 198 normal alleles in this population was estimated to be 2-copy alleles (‘2+0’ genotype). SMN2 copy numbers showed a high degree of individual variation. Our results showed that 64% of the individuals had 2 copies of SMN2, but 36% individuals had between 0, 1, or 3 copies of the gene.
REFERENCES
1.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995. 80:155–65.

2.Ogino S., Wilson RB. Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev Mol Diagn. 2004. 4:15–29.

3.Hendrickson BC., Donohoe C., Akmaev VR., Sugarman EA., Labrousse P., Boguslavskiy L, et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J Med Genet. 2009. 46:641–4.
5.Lee TM., Kim SW., Lee KS., Jin HS., Koo SK., Jo I, et al. Quantitative analysis of SMN1 gene and estimation of SMN1 deletion carrier frequency in Korean population based on real-time PCR. J Korean Med Sci. 2004. 19:870–3.
6.Su YN., Hung CC., Li H., Lee CN., Cheng WF., Tsao PN, et al. Quantitative analysis of SMN1 and SMN2 genes based on DHPLC: a highly efficient and reliable carrier-screening test. Hum Mutat. 2005. 25:460–7.
7.van der Steege G., Grootscholten PM., van der Vlies P., Draaijers TG., Osinga J., Cobben JM, et al. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995. 345:985–6.

8.Wang CC., Chang JG., Jong YJ., Wu SM. Universal multiplex PCR and CE for quantification of SMN1/SMN2 genes in spinal muscular atrophy. Electrophoresis. 2009. 30:1102–10.
9.Scarciolla O., Stuppia L., De Angelis MV., Murru S., Palka C., Giuliani R, et al. Spinal muscular atrophy genotyping by gene dosage using multiple ligation-dependent probe amplification. Neurogenetics. 2006. 7:269–76.

10.Cusin V., Clermont O., Gerard B., Chantereau D., Elion J. Prevalence of SMN1 deletion and duplication in carrier and normal populations: implication for genetic counselling. J Med Genet. 2003. 40:e39.
11.Ogino S., Wilson RB., Gold B. New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur J Hum Genet. 2004. 12:1015–23.
Fig. 1.
Electrophoreogram of SMN1: SMN2=1:3. The carrier is heterozygous for the deletion of the SMN1 gene as shown by reduction in peak heights from 2 probes for exons 7 and 8 (closed arrows). The relative peak height ratios of SMN2 gene increase to approximately 1.5, indicating the presence of 3 copies of SMN2 (open arrows).
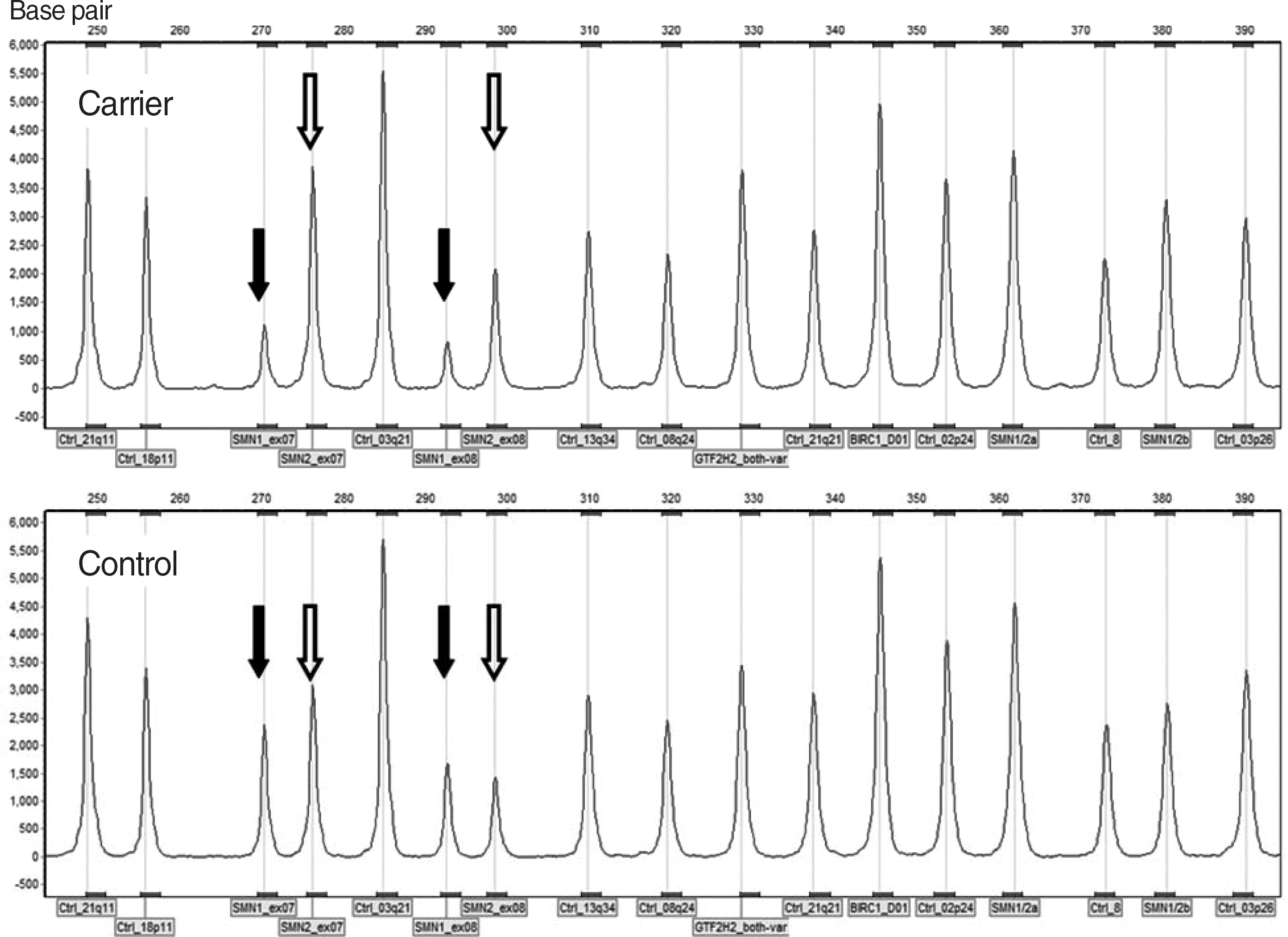
Table 1.
SMN1 and SMN2 copy numbers in a Korean population
| SMN1:SMN2 ratio | No. of subjects | Interpretation |
|---|---|---|
| 1:1 | 1 | SMA carrier |
| 1:3 | 1 | SMA carrier |
| 2:0 | 2 | Noncarrier |
| 2:1 | 28 | Noncarrier |
| 2:2 | 63 | Noncarrier |
| 2:3 | 3 | Noncarrier |
| 3:2 | 1 | Noncarrier |
| 2:2/3:1∗ | 1 | Noncarrier |
| Total | 100 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download