Abstract
A variant Philadelphia chromosome (Ph) is generated from translocation of one or more partner chromosomes in addition to chromosomes 9 and 22. We have described the cases of 2 patients bearing variant Ph detected by interphase FISH but not detected by G-banded karyotyping and metaphase FISH. FISH was performed using BCR/ABL dual color dual fusion translocation probes (Abbott Molecular, USA). A 52-year-old man was diagnosed with acute leukemia of mixed phenotype. G-banded karyotyping showed 46,XY,t(9;22)(q34;q11.2)[12]/47,idem,+der(22)t(9;22)[5]/46,XY[3]. Interphase FISH revealed nuc ish(ABL1,BCR)×3(ABL1 con BCR×2)[329/450]/(ABL1,BCR)×4(ABL1 con BCR×3)[5/450]/(AL1,BCR)×3(ABL1 con BCR×1)[44/450]. Metaphase FISH showed ish (9;22)(ABL1+,BCR1+;BCR+,ABL+)[22]/der(22)(BCR+,ABL1+)[3]. The other case was that of a 31-yr-old male patient diagnosed with CML in the blastic phase. G-banded karyotyping of all 20 metaphase cells showed 47,XYYc,dup(1)(q21q32),del(7)(p11.2),t(9;22)(q34;q11.2). Interphase FISH revealed nuc ish(ABL1,BCR)×3(ABL1 con BCR×2)[254/600]/(ABL1,BCR)×3(ABL1 con BCR×1)[191/600]. Metaphase FISH showed ish t(9;22)(ABL1+,BCR+;BCR+,ABL1+)[16]. These results suggest that typical t(9;22) and variant Ph may coexist in the same patient, and interphase FISH may facilitate the detection of the variant Ph that cannot be detected by G-banded karyotyping alone.
Go to : 
REFERENCES
1.Gorusu M., Benn P., Li Z., Fang M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet Cytogenet. 2007. 173:97–106.

2.So CC., Wan TS., Yip SF., Chan LC. A dual colour dual fusion fluorescence in situ hybridisation study on the genesis of complex variant translocations in chronic myelogenous leukaemia. Oncol Rep. 2008. 19:1181–4.

3.Richebourg S., Eclache V., Perot C., Portnoi MF., Van den Akker J., Terré C, et al. Mechanisms of genesis of variant translocation in chronic myeloid leukemia are not correlated with ABL1 or BCR deletion status or response to imatinib therapy. Cancer Genet Cytogenet. 2008. 182:95–102.
4.Bennour A., Sennana H., Laatiri MA., Elloumi M., Khelif A., Saad A. Molecular cytogenetic characterization of variant Philadelphia translocations in chronic myeloid leukemia: genesis and deletion of derivative chromosome 9. Cancer Genet Cytogenet. 2009. 194:30–7.

5.Mark HF., Sokolic RA., Mark Y. Conventional cytogenetics and FISH in the detection of BCR/ABL fusion in chronic myeloid leukemia (CML). Exp Mol Pathol. 2006. 81:1–7.
6.Shaffer LG., Slovak ML, et al. eds. ISCN. 2009. an international system for human cytogenetic nomenclature (2009). Basel: Karger, 2009.
7.O'Brien S., Thall PF., Siciliano MJ. Cytogenetics of chronic myelogenous leukaemia. Baillieres Clin Haematol. 1997. 10:259–76.
8.El-Zimaity MM., Kantarjian H., Talpaz M., O'Brien S., Giles F., Garcia-Manero G, et al. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br J Haematol. 2004. 125:187–95.

9.Johansson B., Fioretos T., Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002. 107:76–94.

10.Sandberg AA. Chromosomes and causation of human cancer and leukemia: XL. The Ph1 and other translocations in CML. Cancer. 1980. 46:2221–6.

11.Loncarevic IF., Romer J., Starke H., Heller A., Bleck C., Ziegler M, et al. Heterogenic molecular basis for loss of ABL1-BCR transcription: deletions in der(9)t(9;22) and variants of standard t(9;22) in BCR-ABL1-positive chronic myeloid leukemia. Genes Chromosomes Cancer. 2002. 34:193–200.
12.Reid AG., Huntly BJ., Grace C., Green AR., Nacheva EP. Survival implications of molecular heterogeneity in variant Philadelphia-positive chronic myeloid leukaemia. Br J Haematol. 2003. 121:419–27.

13.Yuregir OO., Sahin FI., Yilmaz Z., Kizilkilic E., Karakus S., Ozdogu H. Fluorescent in situ hybridization studies in multiple myeloma. Hematology. 2009. 14:90–4.
14.Chen L., Li J., Xu W., Qiu H., Zhu Y., Zhang Y, et al. Molecular cytogenetic aberrations in patients with multiple myeloma studied by interphase fluorescence in situ hybridization. Exp Oncol. 2007. 29:116–20.
Go to : 
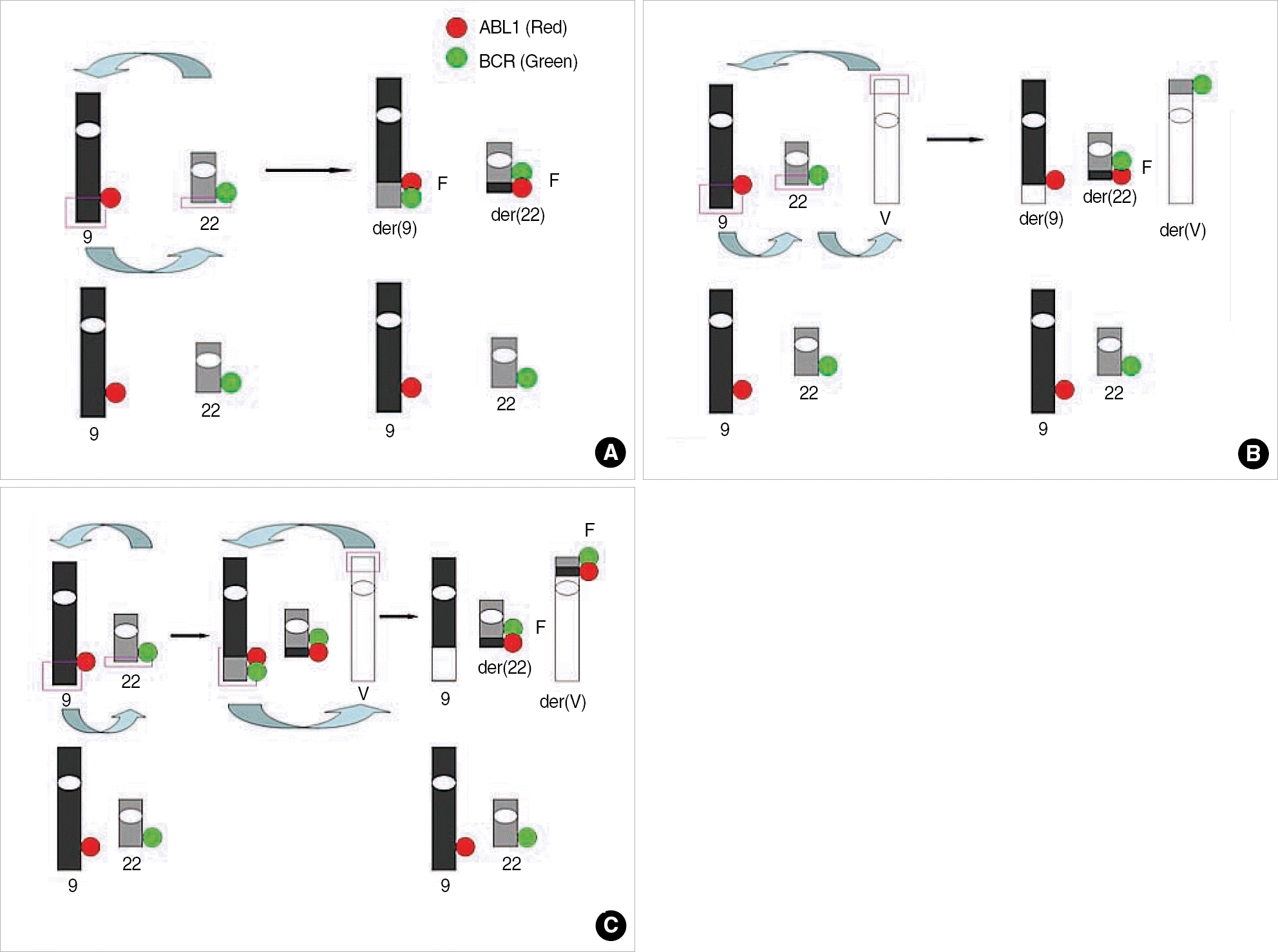 | Fig. 1.Mechanism of detection of classical and variant Philadelphia chromosome (Ph) and signal patterns of FISH using dual color dual fusion probe. (A) Classical translocation showing G1R1F2. (B) One-step mechanism of variant Ph, showing G2R2F1. Three breaks and reciprocal rejoinings occur at the same time on chromosome 9, 22, and partner chromosome. (C) Two-step mechanism of variant Ph, showing G1R1F2. The standard t(9;22) is formed, and then, the segment of der(9) exchanges material with the variable partner chromosome. Modified from figure in reference [1].
Abbreviations: V, variable partner chromosome; G, green signal; R, red signal; F, fusion signal.
|
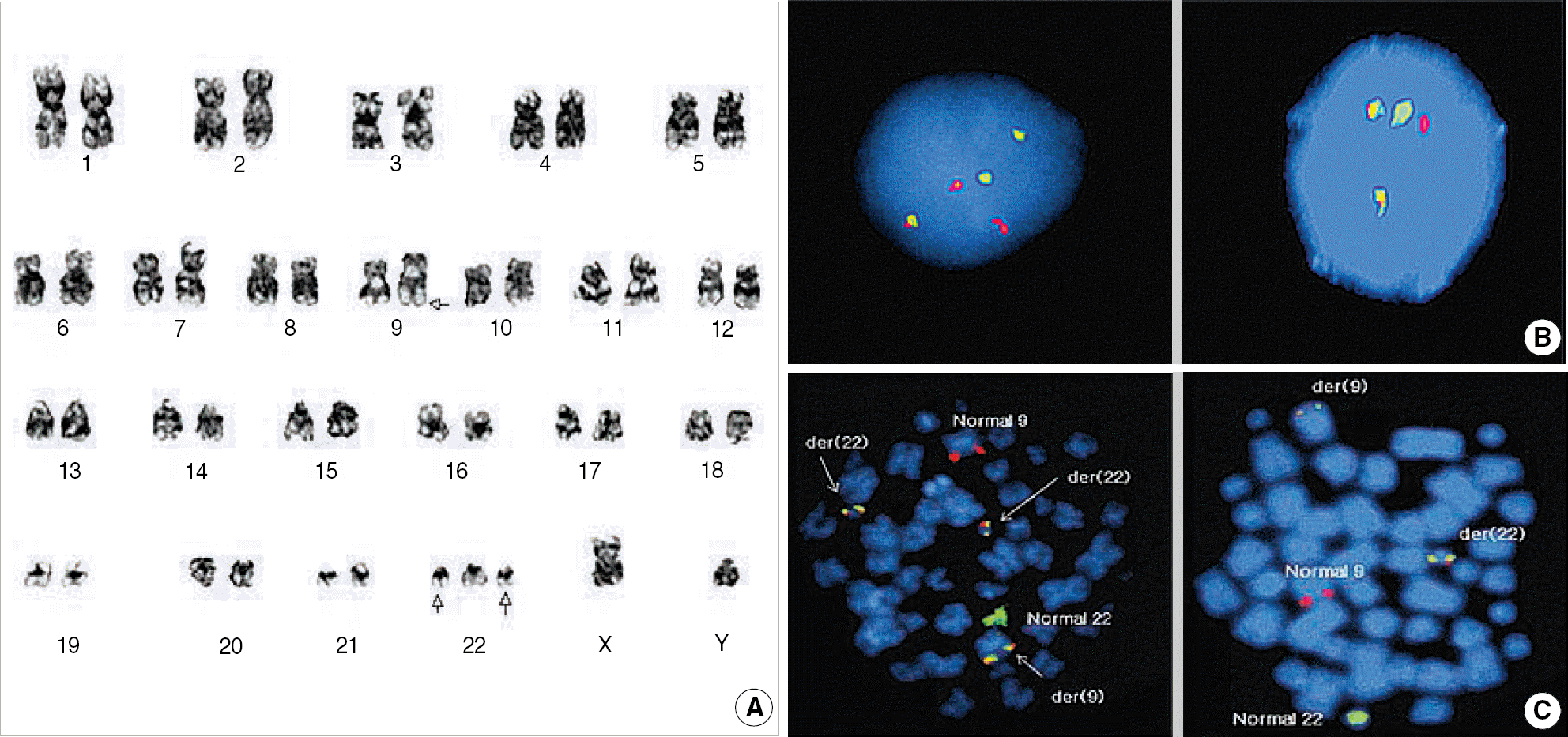 | Fig. 2.Cytogenetic findings of Case 1. (A) G-banded karyogram showing 47,XY,t(9;22)(q34;q11.2),+der(22)t(9;22). (B) Interphase FISH signal pattern showed G2R2F1 (left), suggesting one-step mechanism of variant Ph and G1R1F2 (right), indicating typical t(9;22). (C) Metaphase FISH showed G1R1F3 (left). Each fusion signal resides on the der(22)t(9;22),+der(22)t(9;22), and der(9)t(9;22). G1R1F2 (right) indicate typical t(9;22).
Abbreviations: G, green signal; R, red signal; F, fusion signal.
|
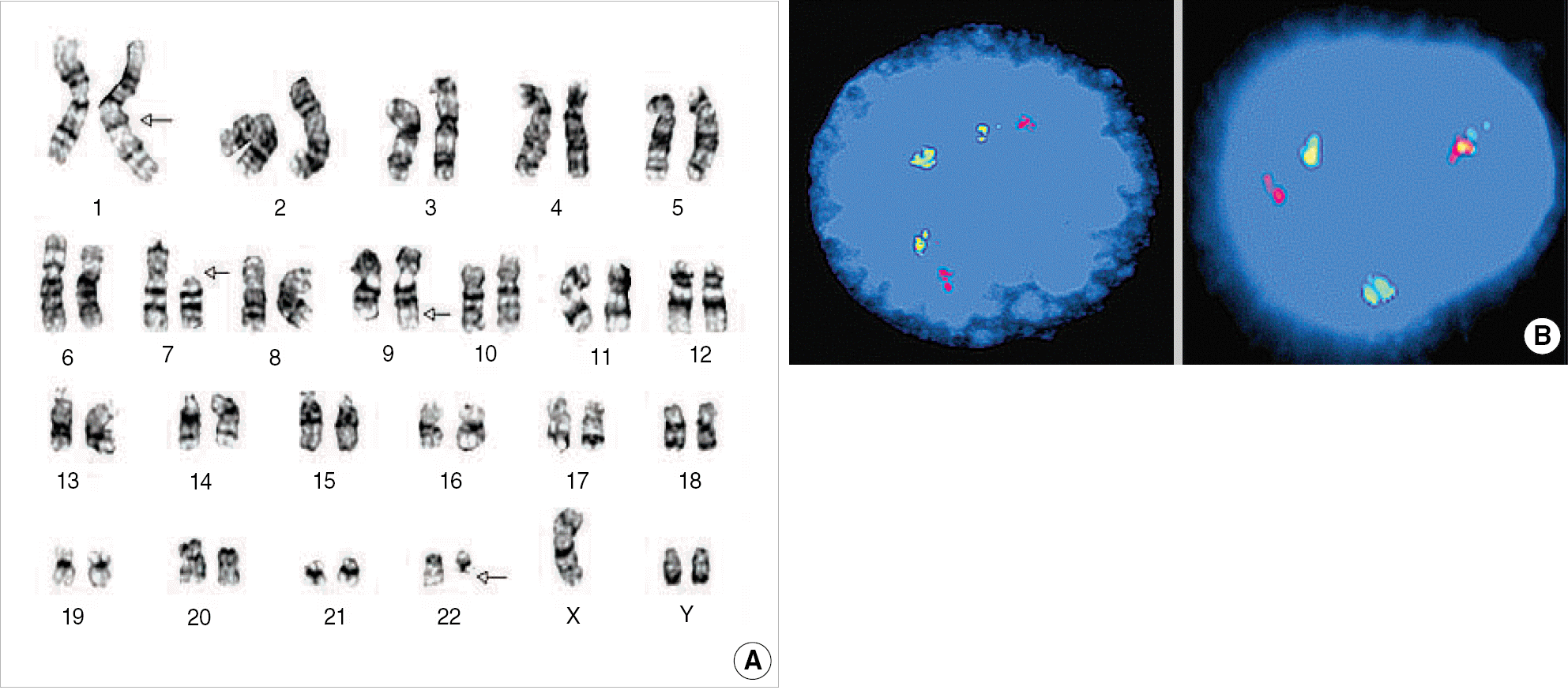 | Fig. 3.Cytogenetic findings of Case 2. (A) G-banded karyogram showing 47,XYYc,dup(1)(q21q32),del(7)(p11.2),t(9;22)(q34;q11.2). (B) Interphase FISH signal pattern of G2R2F1 (left), suggesting one-step mechanism of variant translocation and G1R1F2 (right), indicating typical t(9;22). |
Table 1.
Follow-up data of karyotyping and FISH in case 1
Table 2.
Follow-up data of karyotyping and FISH in case 2




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download