Abstract
Background:
Accurate and early detection of vancomycin-resistant enterococci (VRE) is critical for controlling nosocomial infection. In this study, we evaluated the usefulness of a selective chromogenic agar medium and of multiplex PCR for detection of VRE, and both these techniques were compared with the conventional culture method for VRE detection.
Methods:
We performed the following 3 methods for detecting VRE infection in stool specimens: the routine culture method, culturing in selective chromogenic agar medium (chromID VRE, bioMérieux, France), and multiplex PCR using the Seeplex® VRE ACE Detection kit (Seegene Inc., Korea) with additional PCR for vanC genes.
Results:
We isolated 109 VRE strains from 100 stool specimens by the routine culture method. In chromID VRE, all the isolates showed purple colonies, including Enterococcus gallinarum and E. raffinosus, which were later identified using the Vitek card. All VRE isolates were identified by the multiplex PCR method; 100 were vanA-positive E. faecium, 8 were vanA- and vanC-1-positive E. gallinarum, and 1 was vanA-positive E. raffinosus.
Conclusions:
For VRE surveillance, culturing the isolates in chromID VRE after broth enrichment appears to be an accurate, rapid, and easy method for routine screening test. Multiplex PCR is relatively expensive and needs skilled techniques for detecting VRE, but it can be an auxiliary tool for rapid detection of genotype during a VRE outbreak.
Go to : 
REFERENCES
1.Cuzon G., Naas T., Fortineau N., Nordmann P. Novel chromogenic medium for detection of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. J Clin Microbiol. 2008. 46:2442–4.
2.Song JY., Hwang IS., Eom JS., Cheong HJ., Bae WK., Park YH, et al. Prevalence and molecular epidemiology of vancomycin-resistant enterococci (VRE) strains isolated from animals and humans in Korea. Korean J Intern Med. 2005. 20:55–62.

3.Dutka-Malen S., Evers S., Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995. 33:1434.

4.Delmas J., Robin F., Schweitzer C., Lesens O., Bonnet R. Evaluation of a new chromogenic medium, ChromID VRE, for detection of van-comycin-resistant enterococci in stool samples and rectal swabs. J Clin Microbiol. 2007. 45:2731–3.

5.Lee K., Jang SJ., Lee HJ., Ryoo N., Kim M., Hong SG, et al. Increasing prevalence of vancomycin-resistant Enterococcus faecium, expanded-spectrum cephalosporin-resistant Klebsiella pneumoniae, and imipenem-resistant Pseudomonas aeruginosa in Korea: KONSAR study in 2001. J Korean Med Sci. 2004. 19:8–14.
6.Merquior VL., GonÇalves Neves FP., Ribeiro RL., Duarte RS., de Andrade Marques E., Teixeira LM. Bacteraemia associated with a vancomycin-resistant Enterococcus gallinarum strain harbouring both the vanA and vanC1 genes. J Med Microbiol. 2008. 57:244–5.
7.Grabsch EA., Ghaly-Derias S., Gao W., Howden BP. Comparative study of selective chromogenic (chromID VRE) and bile esculin agars for isolation and identification of vanB-containing vancomycin-resistant enterococci from feces and rectal swabs. J Clin Microbiol. 2008. 46:4034–6.
8.Kuch A., Stefaniuk E., Ozorowski T., Hryniewicz W. New selective and differential chromogenic agar medium, chromID VRE, for screening vancomycin-resistant Enterococcus species. J Microbiol Methods. 2009. 77:124–6.
9.Asir K., Wilkinson K., Perry JD., Reed RH., Gould FK. Evaluation of chromogenic media for the isolation of vancomycin-resistant enterococci from stool samples. Lett Appl Microbiol. 2009. 48:230–3.

10.Kim S., Sung H., Jeon HS., Park SJ., Park SH., Kim MN. Evaluation of a rapid enrichment-PCR method for the detection of vanA vancomycin-resistant enterococci in fecal specimens. Korean J Clin Microbiol. 2007. 10:44–8. (김솔잎, 성흥섭, 전홍선, 박숙자, 박상혁, 김미나.대변검체에서 vanA형 반코마이신내성장구균을검출하기위한신속증균중합효소연쇄반응법평가. 대한임상미생물학회지 2007;10:44-8.).
11.Ledeboer NA., Tibbetts RJ., Dunne WM. A new chromogenic agar medium, chromID VRE, to screen for vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Diagn Microbiol Infect Dis. 2007. 59:477–9.
12.Yoon YK., Sim HS., Kim JY., Park DW., Sohn JW., Roh KH, et al. Epidemiology and control of an outbreak of vancomycin-resistant enterococci in the intensive care units. Yonsei Med J. 2009. 50:637–43.

13.Palladino S., Kay ID., Flexman JP., Boehm I., Costa AM., Lambert EJ, et al. Rapid detection of vanA and vanB genes directly from clinical specimens and enrichment broths by real-time multiplex PCR assay. J Clin Microbiol. 2003. 41:2483–6.
14.Drews SJ., Johnson G., Gharabaghi F., Roscoe M., Matlow A., Tellier R, et al. A 24-hour screening protocol for identification of vancomycin-resistant Enterococcus faecium. J Clin Microbiol. 2006. 44:1578–80.
15.Hsueh PR., Teng LJ., Pan HJ., Chen YC., Wang LH., Chang SC, et al. Emergence of vancomycin-resistant enterococci at a university hospital in Taiwan: persistence of multiple species and multiple clones. Infect Control Hosp Epidemiol. 1999. 20:828–33.

16.Sung HS., Yun KA., Kim MN., Pai CH. An Enterococcus gallinarum strain carrying both vanA and vanC1 genes. Korean J Clin Pathol. 2002. 22:31–3. (성홍섭, 윤경아, 김미나, 배직현. vanA 유전자와 vanC1 유전자를동시에가진 Enterococcus gallinarum 1예. 대한임상병리학회지 2002;22:31-3.).
17.Park IJ., Lee WG., Shin JH., Lee KW., Woo GJ. VanB phenotype-vanA genotype Enterococcus faecium with heterogeneous expression of teicoplanin resistance. J Clin Microbiol. 2008. 46:3091–3.
Go to : 
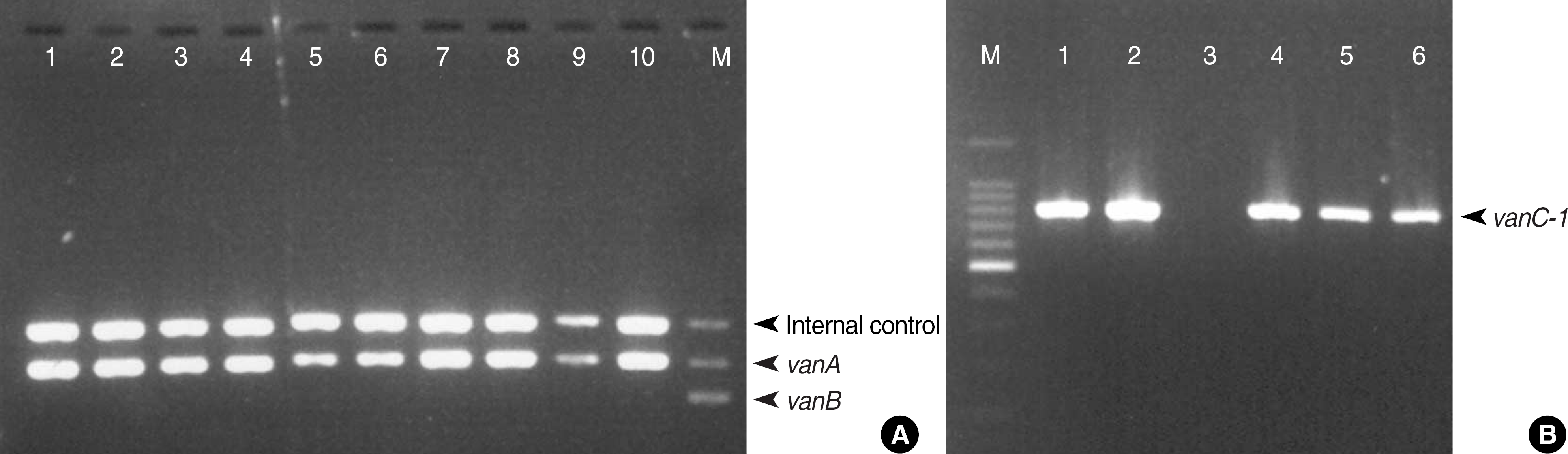 | Fig. 1.Agarose gel electrophoresis of PCR products: Lane M; VRE size marker, Lanes 1, 2, 4-6; E. gallinarum, Lane 3; E. raffinosus, Lanes 7-10; E. faecium. (A) PCR products of clinical isolates of this study using Seeplex® VRE ACE Detection kit: only vanA is observed in all the lanes. (B) PCR products with vanC-1 primer.
Abbreviations: VRE, vancomycin-resistant enterococci; E. gallinarum, Enteroccus gallinarum; E. raffinosus, Enteroccus raffinosus; E. faecium, Enteroccus faecium.
|
Table 1.
Primer sequences that were used in PCR for vancomycin-resistance genotyping
| Amplified gene | Primer sequence (5′-3′) | size (bp) | |
|---|---|---|---|
| vanC-1 | (F) | GGTATCAAGGAAACCTC | 822 |
| (R) | CTTCCGCCATCATAGCT | ||
| vanC-2, vanC-3 | (F) | CTCCTACGATTCTCTTG | 439 |
| (R) | CGAGCAAGACCTTTAAG |
Table 2.
Results of routine culture, chromID, and PCR method of studied isolates
| Routine culture | N of isolates (%) | ChromID (color) | Phenotype (N) | Genotype (by PCR) |
|---|---|---|---|---|
| E. faecium | 100 (91.7) | Purple | VanA (88) VanB (12) | vanA |
| E. gallinarum | 8 (7.3) | Purple | VanA (8) | vanA with vanC-1 |
| E. raffinosus | 1 (0.9) | Purple | VanA (1) | vanA |
| E. casseliflavus 1 (0.9) | NG | NT | NT | |
| Total | 109∗ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download