Abstract
Background:
Although the pericentric inversion of chromosome 9, inv(9)(p11q13), is generally considered a normal variation, it is also associated with solid tumors and several hematologic malignancies such as biphenotypic acute leukemia, ALL, AML, and myeloproliferative neoplasms. However, to the best of our knowledge, there have been no reports that suggest an association between CML and constitutional pericentric inversion of chromosome 9. The purpose of this retrospective study was to investigate the frequency and clinical features of CML patients with concomitant inv(9) and t(9;22)(q34;q11.2) variation at our institution.
Methods:
We reviewed the bone marrow chromosome database entries between October 2006 and December 2008 to identify patients with concomitant inv(9) and t(9;22) variations. Laboratory and clinical data of the patients were obtained from the electronic medical record system.
Conclusions:
Although the association between inv(9) variation and CML is still controversial, we believe that hematologists should consider the role of constitutional inv(9) variation in CML patients to avoid overlooking the impaired engraftment potential of hematopoietic stem cells harboring inv(9). Therefore, we suggest that more effort should be invested to develop cytogenetic tests for detecting constitutional inv(9) variation in CML patients.
REFERENCES
1.Keung YK., Knovich MA., Powell BL., Buss DH., Pettenati M. Constitutional pericentric inversion of chromosome 9 and acute leukemia. Cancer Genet Cytogenet. 2003. 145:82–5.
2.Huh J., Chung W. Incidence and types of constitutional chromosomal abnormalities in patients with hematologic malignancies. Korean J Lab Med. 2006. 26:64–9.

3.Mitelman F., Johansson B., Mertens F. Mitelman database of chromosome aberrations in cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman. (Updated on Nov. 2008.
4.Betz JL., Behairy AS., Rabionet P., Tirtorahardjo B., Moore MW., Cotter PD. Acquired inv(9): what is its significance? Cancer Genet Cytogenet. 2005. 160:76–8.

5.Wan TS., Ma SK., Chan LC. Acquired pericentric inversion of chromosome 9 in essential thrombocythemia. Hum Genet. 2000. 106:669–70.

6.Alimena G., Billstrom R., Casalone R., Gallo E., Mitelman F., Pasquali F. Cytogenetic pattern in leukemic cells of patients with constitutional chromosome anomalies. Cancer Genet Cytogenet. 1985. 16:207–18.

7.Benitez J., Valcarcel E., Ramos C., Ayuso C., Cascos AS. Frequency of constitutional chromosome alterations in patients with hematologic neoplasias. Cancer Genet Cytogenet. 1987. 24:345–54.

8.Cerretini R., Acevedo S., Chena C., Belli C., Larripa I., Slavutsky I. Evaluation of constitutional chromosome aberrations in hematologic disorders. Cancer Genet Cytogenet. 2002. 134:133–7.

9.Welborn J. Constitutional chromosome aberrations as pathogenetic events in hematologic malignancies. Cancer Genet Cytogenet. 2004. 149:137–53.

10.Imashuku S., Naya M., An B., Nakabayashi Y., Kuriyama K., Udeda I, et al. Constitutional pericentric inversion of chromosome 9 and haemopoietic stem cell transplantation: delayed engraftment. Br J Haematol. 2002. 118:1195–6.

11.Keung YK., Knovich MA., Hurd DD., Pettenati M. Constitutional pericentric inversion of chromosome 9 and bone marrow transplantation. Br J Haematol. 2003. 123:748–9.

12.Bernstein R., Pinto MR., Wallace C., Penfold G., Mendelow B. The incidence, type, and subsequent evolution of 14 variant Ph1 trans-locations in 180 South African patients with Ph1-positive chronic myeloid leukemia. Cancer Genet Cytogenet. 1984. 12:225–38.

13.Lee DS., Lee YS., Yun YS., Kim YR., Jeong SS., Lee YK, et al. A study on the incidence of ABL gene deletion on derivative chromosome 9 in chronic myelogenous leukemia by interphase fluorescence in situ hybridization and its association with disease progression. Genes Chromosomes Cancer. 2003. 37:291–9.
14.Lim LC., Heng KK., Vellupillai M., Tan LT., Boey BC., Lau LC, et al. Molecular and phenotypic spectrum of de novo Philadelphia positive acute leukemia. Int J Mol Med. 1999. 4:665–7.

15.Lo Nigro L., Sainati L., Mirabile E., Lanciotti M., Poli A., Leszl A, et al. Association of cytogenetic abnormalities with detection of BCR-ABL fusion transcripts in children with T-lineage lymphoproliferative diseases (T-ALL and T-NHL). Pediatr Blood Cancer. 2004. 42:278–80.
16.Ohtaki K., Abe R., Tebbi CK., de los Santos R., Han T., Sandberg AA. Near-triploid Ph-positive leukemia. Cancer Genet Cytogenet. 1985. 18:113–21.

17.Peterson LC., Parkin JL., Arthur DC., Brunning RD. Acute basophilic leukemia. A clinical, morphologic, and cytogenetic study of eight cases. Am J Clin Pathol. 1991. 96:160–70.

18.Lourenco GJ., Silva PM., Bognone RA., De Souza RA., Delamain MT., Lima CS. Inherited pericentric inversion of chromosome 9 in acquired hematological disorders. Ann Hematol. 2007. 86:465–7.

Fig. 1.
The full karyogram of bone marrow cells of patient 1 is as follows: 46,XY,inv(9)(p11q13)c,t(9;22)(q34;q11.2). The bold arrow denotes inv(9)(p11q13) and the single arrows denote t(9;22)(q34;q11.2).
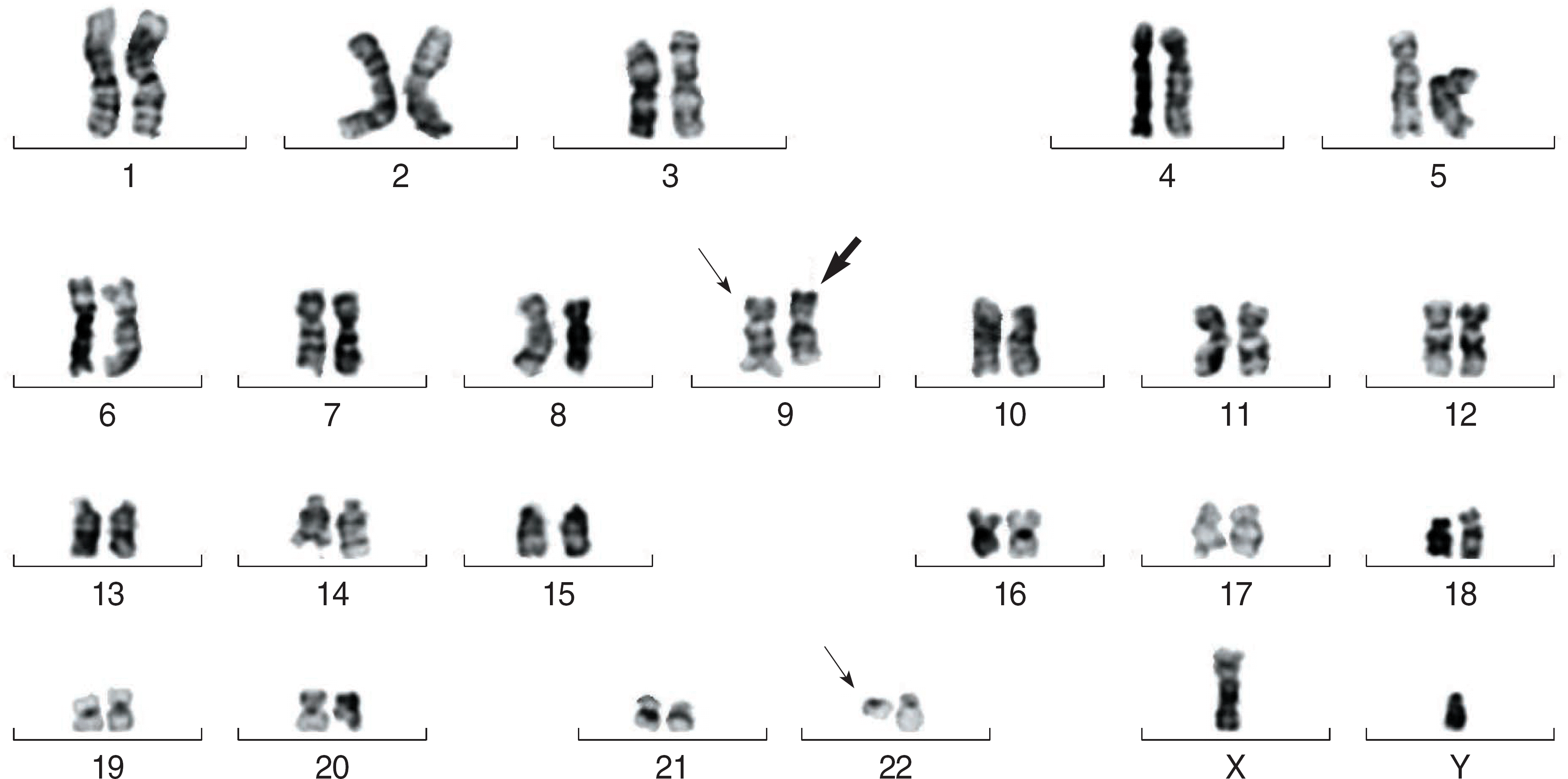
Fig. 2.
Multiplex reverse transcriptase-PCR (RT-PCR) was performed using Seeplex Leukemia BCR/ABL kit (Seegene, Seoul, Korea). Lane M, molecular marker provided by manufacturer; Lane 1, sample from Patient 1 (b3a2 type); Lane 2, negative control; Lane 3, sample from Patient 3 (b2a2 type); Lane 4, positive control (e1a2 type); Lane 5, RT-PCR product of b2a2/e1a2 as a positive control; Lane 6, blank; Lane L, molecular size marker (100-bp ladder). Target and amplicon sizes are as follows: internal control (600 bp), b3a2 (476 bp), b2a2 (401 bp), and e1a2 (348 bp).
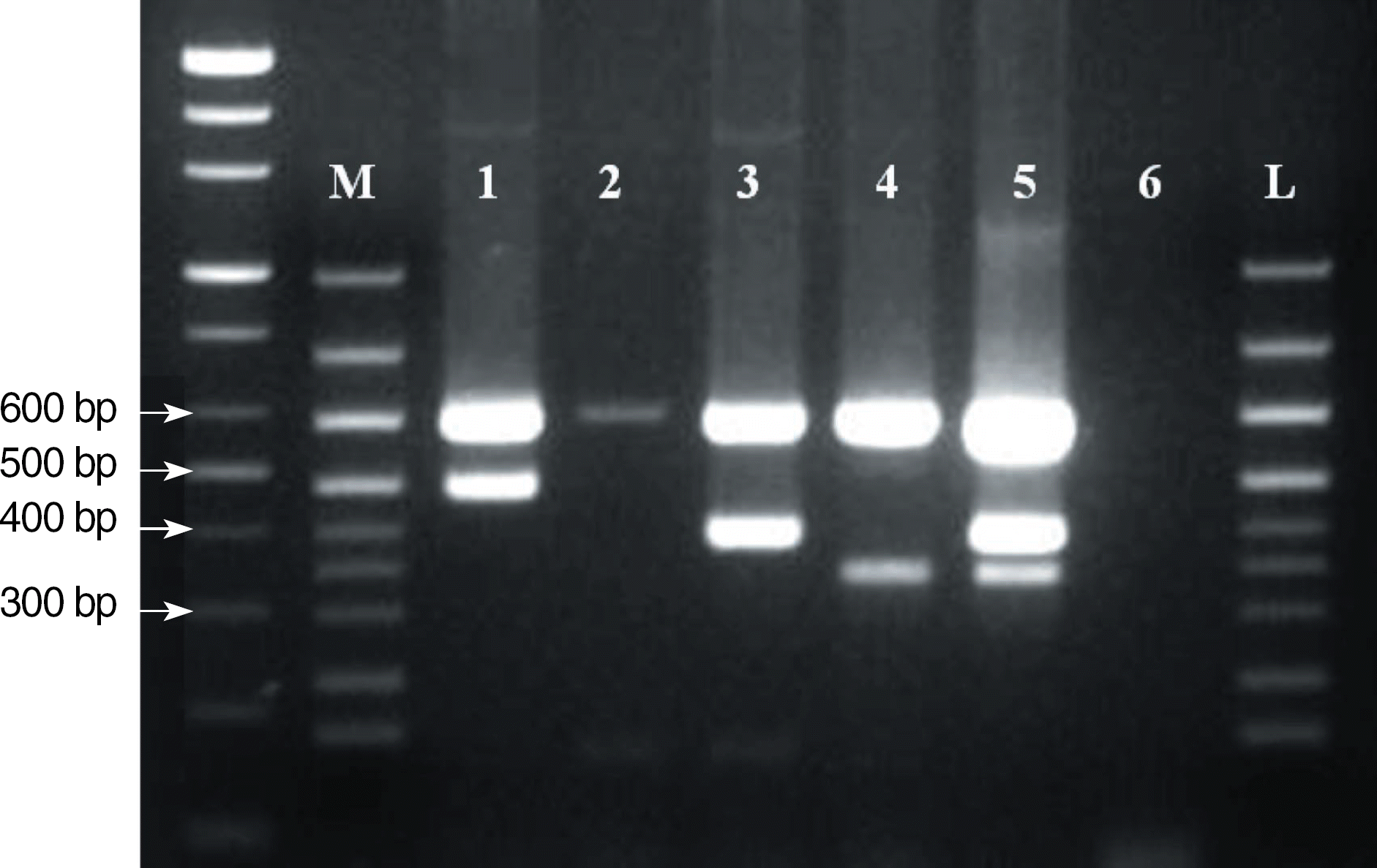
Fig. 3.
The partial karyogram of bone marrow cells of patients 2, 3, and 4. The bold arrows denote inv(9)(p11q13) and the single arrows denote t(9;22)(q34;q11.2).
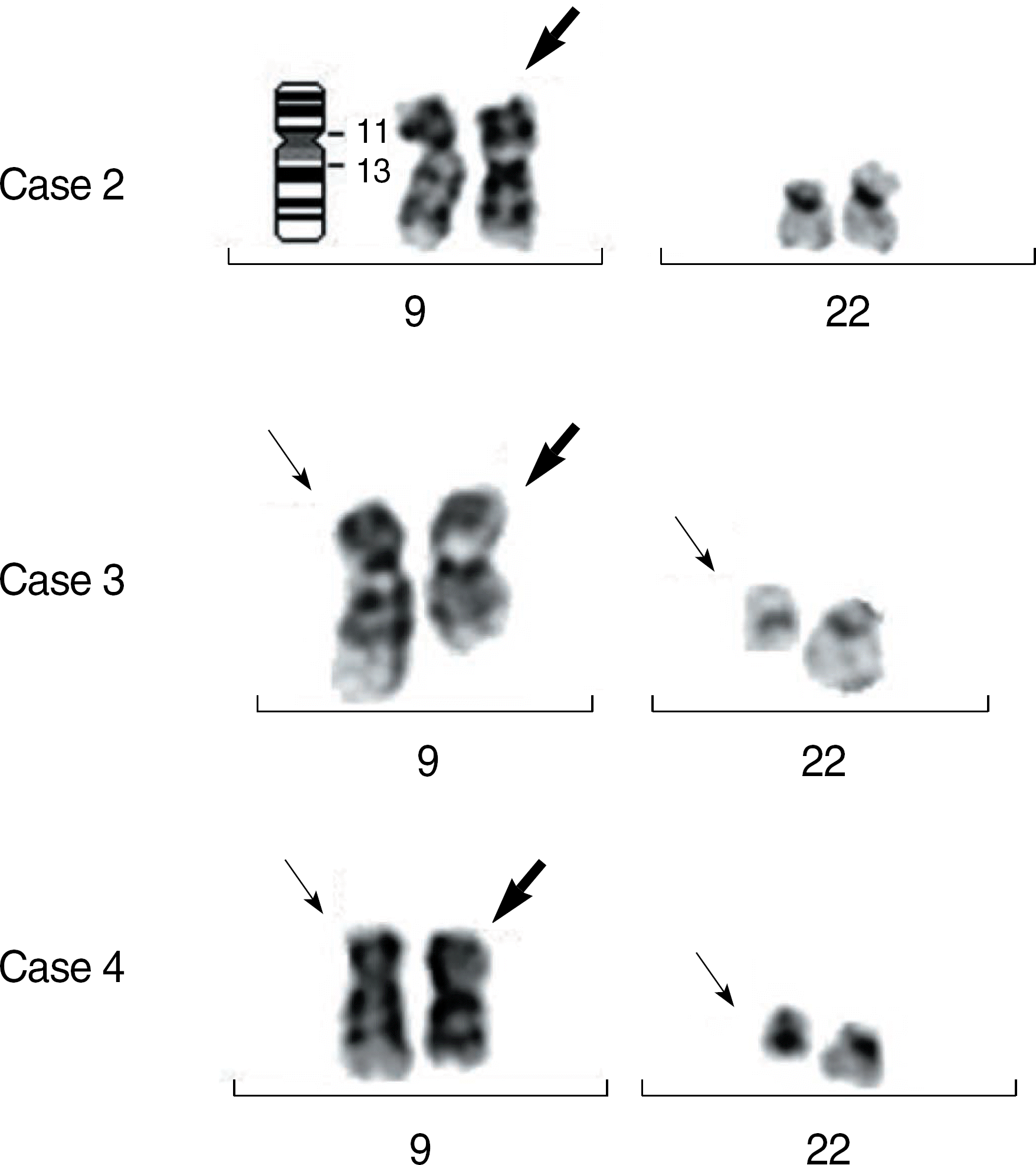
Table 1.
Reported cases of concomitant inv(9) and t(9;22) variations in Ph+ leukemia patients




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download