Abstract
Background:
Neurofibromatosis type 2 (NF2) is an autosomal dominant syndrome caused by the NF2 tumor suppressor gene. However, the NF2 mutation characteristics in Korean patients are not sufficiently understood. In this study, we conducted a comprehensive mutational analysis in 7 Korean NF2 patients by performing direct sequencing and gene-dosage assessment.
Methods:
We analyzed all exons and flanking regions of NF2 by direct sequencing and screened the deletions or duplications involving NF2 by multiplex ligation-dependent probe amplification.
Results:
Four novel NF2 mutations, including 2 splice-site mutations (c.364-1G>A and c.886-3C>G), 1 frameshift mutation (c.524delA), and 1 missense mutation (c.397T>C; p.Cys133Arg), were identified in our patients. No large deletion or duplication was identified in our series. Subsequently, we identified an abnormal splicing product by using reverse transcription-PCR and direct sequencing in 2 patients with a novel splice-site mutation. The missense mutation c.397T>C was predicted to have harmful effects on protein function.
REFERENCES
1.Evans DG., Huson SM., Donnai D., Neary W., Blair V., Newton V, et al. A clinical study of type 2 neurofibromatosis. Q J Med. 1992. 84:603–18.
2.Wallace AJ., Watson CJ., Oward E., Evans DG., Elles RG. Mutation scanning of the NF2 gene: an improved service based on meta-PCR/sequencing, dosage analysis, and loss of heterozygosity analysis. Genet Test. 2004. 8:368–80.
3.Kluwe L., Nygren AO., Errami A., Heinrich B., Matthies C., Tatagiba M, et al. Screening for large mutations of the NF2 gene. Genes Chromosomes Cancer. 2005. 42:384–91.
4.Yang HJ., Won YJ., Park KJ., Jung HW., Choi KS., Park JG. Germline mutations of the NF2 gene in Korean neurofibromatosis 2 patient. J Korean Cancer Assoc. 1998. 30:790–9.
5.Ferrer-Costa C., Orozco M., de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004. 57:811–9.

7.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002. 30:3894–900.

8.Ahronowitz I., Xin W., Kiely R., Sims K., MacCollin M., Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007. 28:1–12.
Fig. 1.
Results of RT-PCR and sequence analysis for the 2 splice-site mutations c.364-1G>A (patient P4) and c.886-3C>G (patient P2). (A) The c.364-1G>A mutation yields a 0.55-kb abnormal product as well as 0.63-kb normal product in RT-PCR with the following primers: F-5′-AAGCAACCCAAGACGTTCAC-3′, R-5′-CCGGATTGCAAAGTAGTTCA-3′. (B) The c.886-3C>G cannot be discriminated from normal products by using following primers: F-5′-CTGACCCCCAAGATCTCCT-3′, R-5′-GCTTCAGCTGATCTGCCTCT-3′. (C) Exon 4 skipping is shown in the cDNA sequence of patient 4 with c.364-1G>A. (D) Patient P2 is heterozygous for c.886-3C>G, and 2-bp insertion of AG (blue) between exon 9 (yellow) and exon 10 (pink) is shown in the cDNA sequence. This insertion is caused by the introduction of a new splice acceptor site from c.886-4A to c.886-3C>G and subsequent inclusion of original splice acceptor site (c.886-2_-1AG) in the mature transcript.
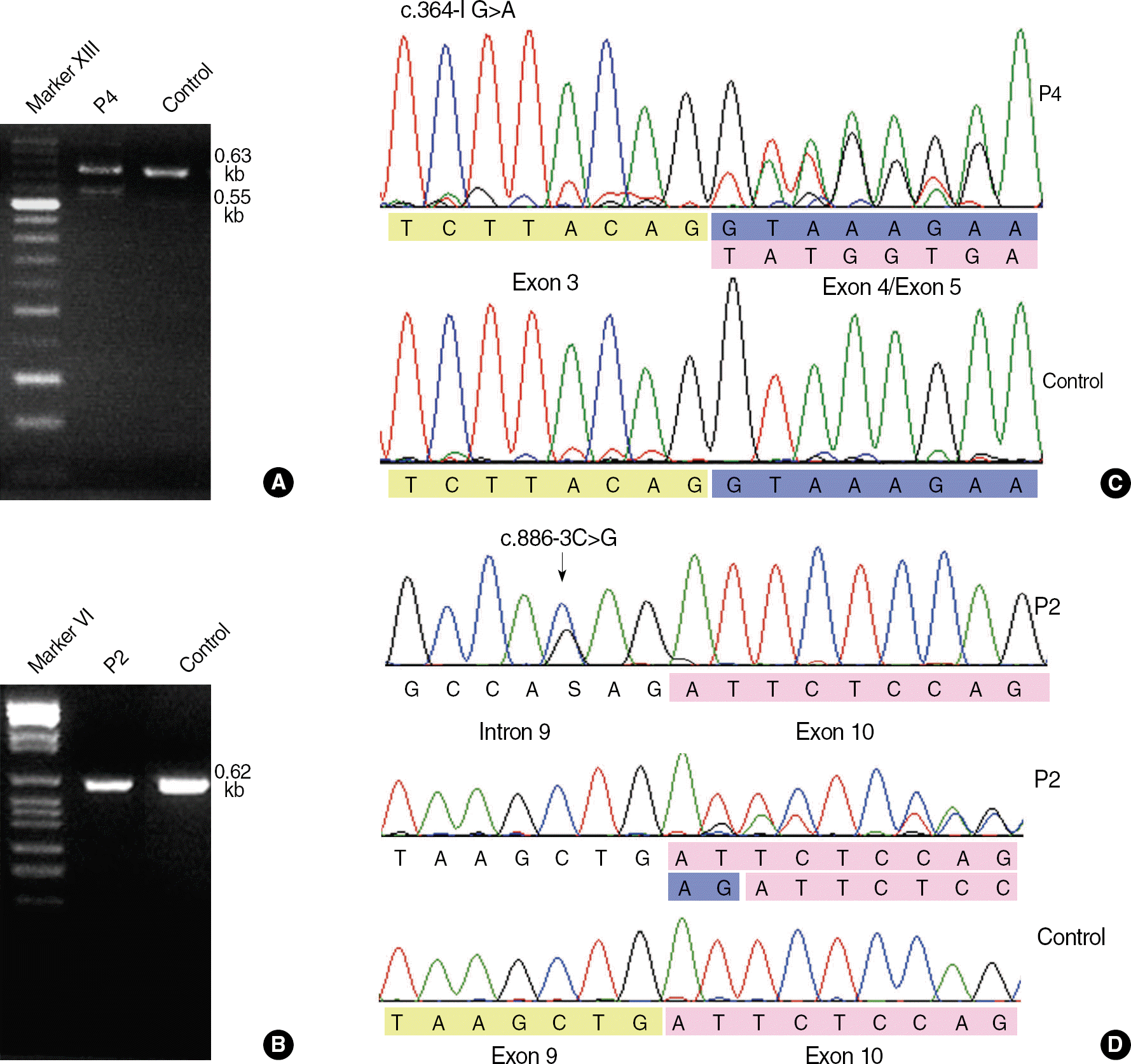
Fig. 2.
Multiple alignment and amino acid conservation for a novel missense mutation c.397T>C (p.Cys133Arg). The cysteine at codon 133 is well-conserved among various species.

Table 1.
Manchester clinical diagnostic criteria for neurofibromatosis type 2
Table 2.
Primer sequences used in this study
Table 3.
Clinical features and molecular findings of the 7 patients who participated in this study




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download