Abstract
Background
Microdeletion syndromes not detectable by conventional cytogenetic analysis have been reported to occur in approximately 5% of patients with unexplained mental retardation (MR). Therefore, it is essential to ensure that patients with MR are screened for these microdeletion syndromes. Mental retardation syndrome multiplex ligation-dependent probe amplification (MRS-MLPA) is a new technique for measuring sequence dosages that allows for the detection of copy number changes of several microdeletion syndromes (1p36 deletion syndrome, Williams syndrome, Smith-Magenis syndrome, Miller-Dieker syndrome, DiGeorge syndrome, Prader-Willi/Angelman syndrome, Alagille syndrome, Saethre-Chotzen syndrome, and Sotos syndrome) to be processed simultaneously, thus significantly reducing the amount of laboratory work.
Methods
We assessed the performance of MLPA (MRC-Holland, The Netherlands) for the detection of microdeletion syndromes by comparing the results with those generated using FISH assays. MLPA analysis was carried out on 12 patients with microdeletion confirmed by FISH (three DiGeorge syndrome, four Williams syndrome, four Prader-Willi syndrome, and one Miller-Dieker syndrome).
Go to : 
REFERENCES
1.Kirchhoff M., Bisgaard AM., Bryndorf T., Gerdes T. MLPA analysis for a panel of syndromes with mental retardation reveals imbalances in 5.8% of patients with mental retardation and dysmorphic features, including duplications of the Sotos syndrome and Williams-Beuren syndrome regions. Eur J Med Genet. 2007. 50:33–42.

2.Hunter AG. Outcome of the routine assessment of patients with mental retardation in a genetics clinic. Am J Med Genet. 2000. 90:60–8.

3.Sellner LN., Taylor GR. MLPA and MAPH: new techniques for detection of gene deletions. Hum Mutat. 2004. 23:413–9.

4.Rusu C., Sireteanu A., Puiu M., Skrypnyk C., Tomescu E., Csep K, et al. MLPA technique-principles and use in practice. Rev Med Chir Soc Med Nat Iasi. 2007. 111:1001–4.
5.Rauch A., Hoyer J., Guth S., Zweier C., Kraus C., Becker C, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006. 140:2063–74.

6.Flint J., Knight S. The use of telomere probes to investigate sub-microscopic rearrangements associated with mental retardation. Curr Opin Genet Dev. 2003. 13:310–6.

7.Shaffer LG., Bejjani BA. A cytogeneticist's perspective on genomic microarrays. Hum Reprod Update. 2004. 10:221–6.

8.Wehner M., Mangold E., Sengteller M., Friedrichs N., Aretz S., Friedl W, et al. Hereditary nonpolyposis colorectal cancer: pitfalls in deletion screening in MSH2 and MLH1 genes. Eur J Hum Genet. 2005. 13:983–6.

9.Armour JA., Palla R., Zeeuwen PL., den Heijer M., Schalkwijk J., Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007. 35:e19.

10.White SJ., Vissers LE., Geurts van Kessel A., de Menezes RX., Kalay E., Lehesjoki AE, et al. Variation of CNV distribution in five different ethnic populations. Cytogenetic Genome Res. 2007. 118:19–30.

11.Janssen B., Hartmann C., Scholz V., Jauch A., Zschocke J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005. 6:29–35.

Go to : 
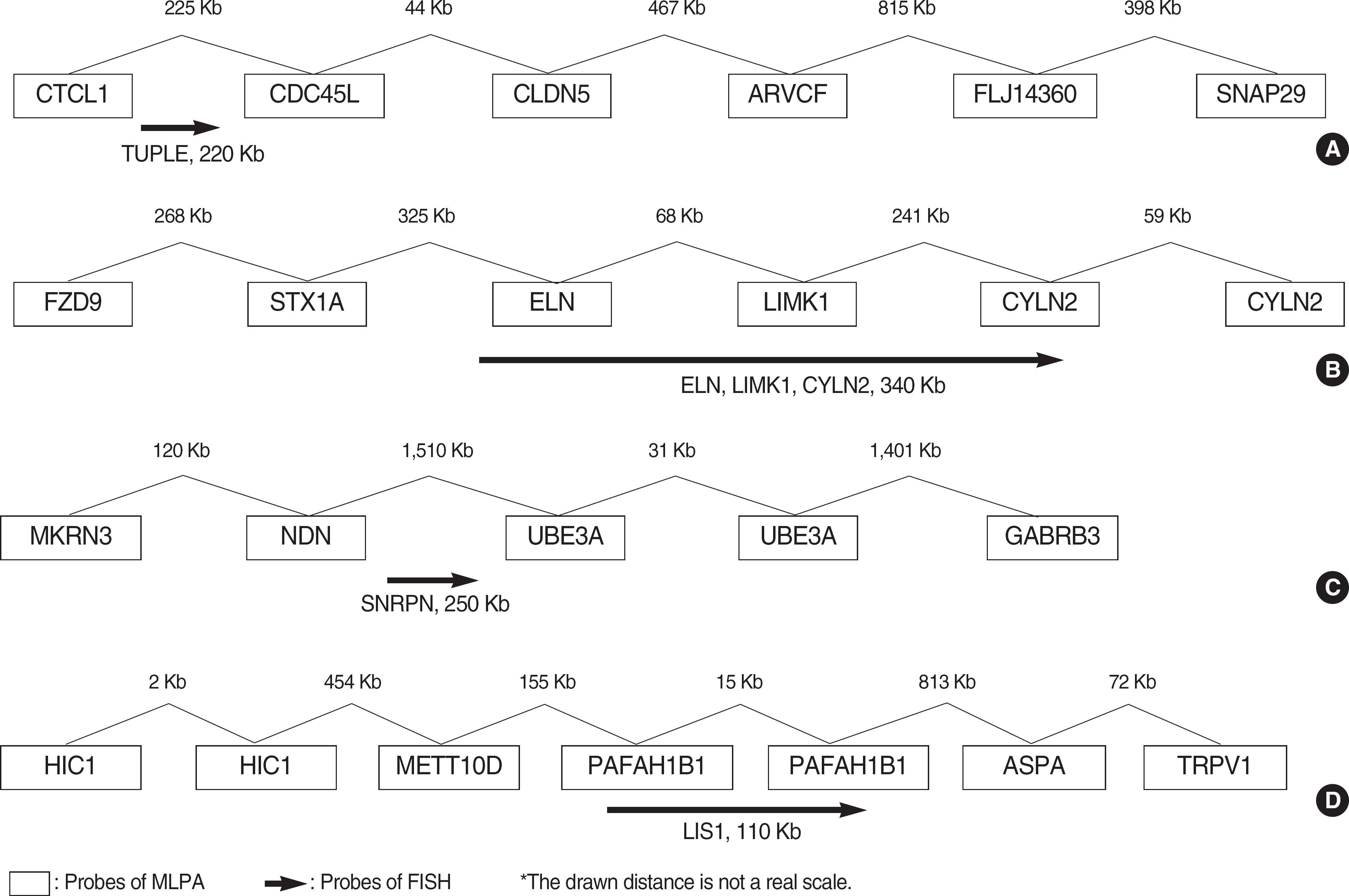 | Fig. 1.The positions of FISH probes compared to MLPA probes. (A) DiGeorge syndrome, (B) Williams syndrome, (C) Prader-Willi/Angelman syndrome, (D) Miller-Dieker syndrome. Abbreviation: MLPA, multiplex ligand-dependent probe amplification. |
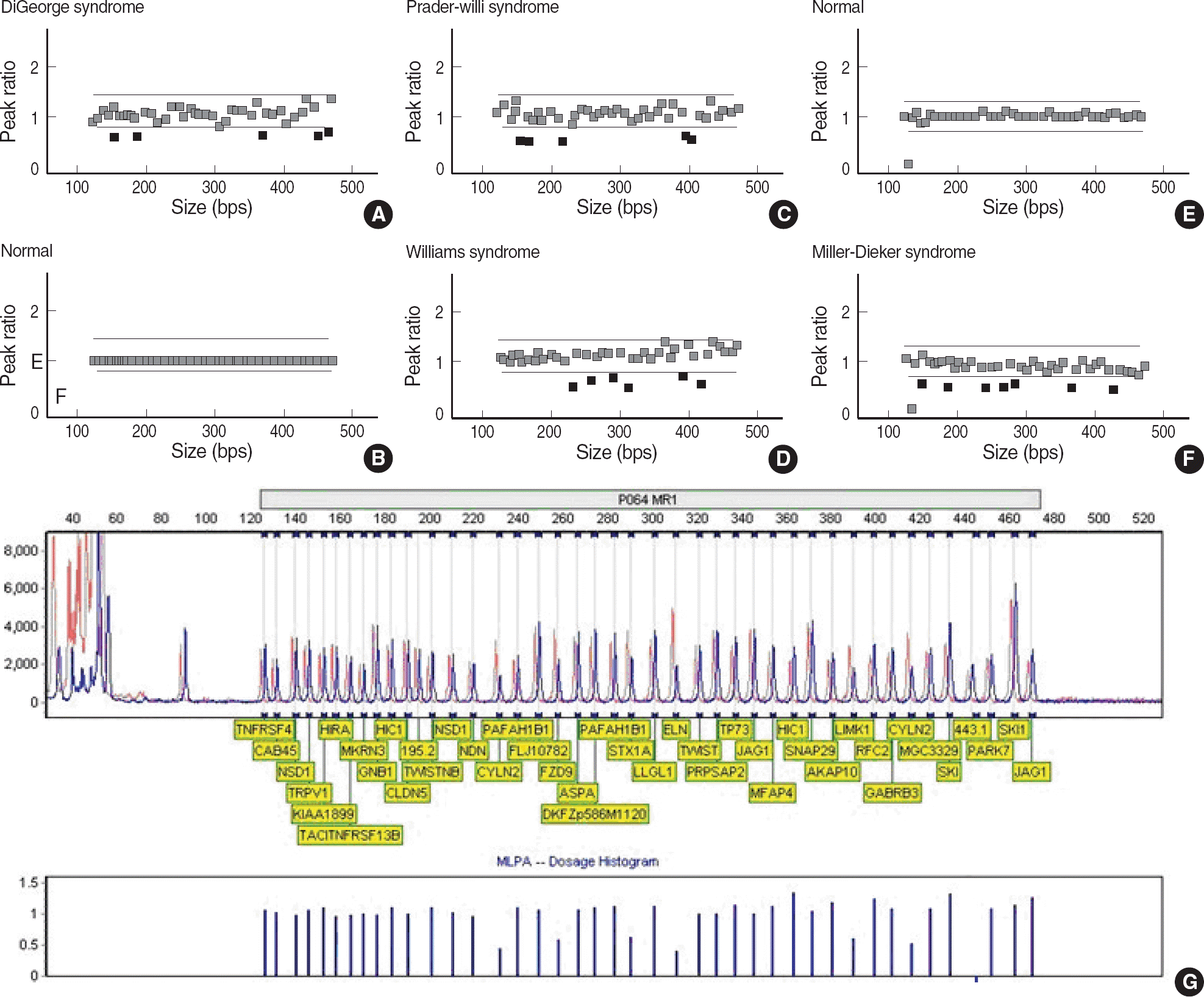 | Fig. 2.MLPA results were analyzed employing Genemarker version 1.6 software. The red dot indicates a probe of peak ratio less than 0.75 compared with normal control. The green dot indicates a probe of peak ratio 0.75-1.3 compared with normal control. (A) MLPA results of a patient with DiGeorge syndrome showed the deletions in five probes for CTCL1, CDC45L, CLDN5, ARVCF, and FLJ14360 genes. (B) MLPA results of normal control. (C) MLPA results of a patient with Prader-Willi/Angelman syndrome showed the deletions in five probes of MKRN3, NDN, UBE3A, and GABRB3 genes. (D) MLPA results of a patient with Williams syndrome showed the deletions in six probes of FZD9, STX1A, ELN, LIMK1, and CYLN2 genes. (E) MLPA results of normal control. (F) MLPA results of Miller-Dieker syndrome showed the deletions in seven probes of HIC1, METT10D, PAFAH1B1, ASPA, and TRPV1 genes. (G) MLPA dosage histograms of Williams syndrome. Abbreviation: MLPA, multiplex ligation-dependent probe amplification. |
Table 1.
Characteristics of MLPA probes used for detection of several microdeletion syndromes




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download