Abstract
Background:
Cytogenetic abnormalities are one of the most reliable prognostic factors in acute leukemia. Combination of conventional chromosome analysis (CCA) and FISH provides higher sensitivity in detecting these genetic abnormalities, and it is effective to apply several FISH probes as a profile test. The objective of this study was to investigate the utility of FISH profile analyses in the initial diagnosis of acute leukemia.
Methods:
Two hundred and forty one de novo acute leukemia patients diagnosed from January, 2002 to November, 2007 were included. For acute lymphoblastic leukemia profile test, FISH probes for BCR/ABL, TEL/AML1, MLL gene rearrangement and CDKN2A deletion were used. For acute myeloid leukemia profile test, probes for AML1/ETO, MLL and CBFβ gene rearrangement were used. The results of CCA and FISH profile tests were collected, and the positive rates were compared.
Results:
ALL FISH profile tests revealed additional genetic aberrations not detected by chromosome analysis in 48.6% (67/138) of cases, including those with normal karyotypes or no mitotic cells (37%, 51/138). Among these 51 cases, TEL/AML1 abnormalities were detected in 44.3%, followed by the abnormal CDKN2A signal (24.6%) and hyperdiploidy (18.0%). AML FISH profile tests revealed additional genetic abnormalities in 7.8% (8/103) of cases.
Conclusions:
FISH analysis as a profile test detected additional genetic aberrations in a significant proportion of acute leukemia, and was effective especially in detecting cryptic translocations, submicroscopic deletions and complex karyotypes. Our study supports the need to incorporate FISH profile test at initial work up in acute leukemia.
REFERENCES
1.Micallef-Eynaud PD., Eden OB., Grace E., Ellis PM. Cytogenetic abnormalities in childhood acute lymphoblastic leukemia. Pediatr Hematol Oncol. 1993. 10:25–30.

2.Faderl S., Kantarjian HM., Talpaz M., Estrov Z. Clinical significance of cytogenetic abnormalities in adult acute lymphoblastic leukemia. Blood. 1998. 91:3995–4019.

3.Grimwade D., Walker H., Oliver F., Wheatley K., Harrison C., Harrison G, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998. 92:2322–33.
4.Slovak ML., Kopecky KJ., Cassileth PA., Harrington DH., Theil KS., Mohamed A, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood. 2000. 96:4075–83.

5.Grimwade D., Walker H., Harrison G., Oliver F., Chatters S., Harrison CJ, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001. 98:1312–20.

6.Ribera JM., Ortega JJ., Oriol A., Granada I., Hernandez-Rivas JM., Parody R, et al. Prognostic value of karyotypic analysis in children and adults with high-risk acute lymphoblastic leukemia included in the PETHEMA ALL-93 trial. Haematologica. 2002. 87:154–66.
7.McGrattan P., Campbell S., Cuthbert R., Jones FG., McMullin MF., Humphreys M. Integration of conventional cytogenetics, comparative genomic hybridisation and interphase fluorescence in situ hybridisation for the detection of genomic rearrangements in acute leukaemia. J Clin Pathol. 2008. 61:903–8.

8.Cox MC., Panetta P., Venditti A., Del Poeta G., Franchi A., Buccisano F, et al. Comparison between conventional banding analysis and FISH screening with an AML-specific set of probes in 260 patients. Hematol J. 2003. 4:263–70.

9.Wolff DJ., Bagg A., Cooley LD., Dewald GW., Hirsch BA., Jacky PB, et al. Guidance for fluorescence in situ hybridization testing in hematologic disorders. J Mol Diagn. 2007. 9:134–43.

10.Sreekantaiah C. FISH panels for hematologic malignancies. Cytogenet Genome Res. 2007. 118:284–96.

11.Jaffe ES, Harris NL, editors. World Health Organization classification of tumours: pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press;2001.
12.Mitelman F, editor. An international system for human cytogenetic nomenclature. Basel: S Karger;1995.
13.Shaffer LG, Tommerup N, editors. An international system for human cytogenetic nomenclature. Basel: S Karger;2005.
14.Andreasson P., Hoglund M., Bekassy AN., Garwicz S., Heldrup J., Mitelman F, et al. Cytogenetic and FISH studies of a single center consecutive series of 152 childhood acute lymphoblastic leukemias. Eur J Haematol. 2000. 65:40–51.

15.Perez-Vera P., Salas C., Montero-Ruiz O., Frias S., Dehesa G., Jarquin B, et al. Analysis of gene rearrangements using a fluorescence in situ hybridization method in Mexican patients with acute lymphoblastic leukemia: experience at a single institution. Cancer Genet Cytogenet. 2008. 184:94–8.
16.Alvarez Y., Gaitan S., Perez A., Bastida P., Ortega JJ., Dastugue N, et al. ETV6/RUNX1 rearrangement in childhood B-precursor acute lymphoblastic leukemia with normal karyotypes or without cytogenetic results. Cancer Genet Cytogenet. 2004. 152:77–80.
17.Loh ML., Silverman LB., Young ML., Neuberg D., Golub TR., Sallan SE, et al. Incidence of TEL/AML1 fusion in children with relapsed acute lymphoblastic leukemia. Blood. 1998. 92:4792–7.
18.Kempski H., Chalker J., Chessells J., Sturt N., Brickell P., Webb J, et al. An investigation of the t(12;21) rearrangement in children with B-precursor acute lymphoblastic leukaemia using cytogenetic and molecular methods. Br J Haematol. 1999. 105:684–9.

19.Cuneo A., Bigoni R., Cavazzini F., Bardi A., Roberti MG., Agostini P, et al. Incidence and significance of cryptic chromosome aberrations detected by fluorescence in situ hybridization in acute myeloid leukemia with normal karyotype. Leukemia. 2002. 16:1745–51.

20.Zhang J., Liu Z., Shao H., Ma Y., Tong H., Wang Y. Laboratory study of a complex translocation t(2;8;21) (p12;q22;q22) in a patient with acute myelogenous leukemia. Leuk Lymphoma. 2008. 49:1925–8.
21.Miyamoto T., Nagafuji K., Akashi K., Harada M., Kyo T., Akashi T, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996. 87:4789–96.
22.Cho EK., Bang SM., Ahn JY., Yoo SM., Park PW., Seo YH, et al. Prognostic value of AML 1/ETO fusion transcripts in patients with acute myelogenous leukemia. Korean J Intern Med. 2003. 18:13–20.
23.Litmanovich D., Zamir-Brill R., Jeison M., Gershoni-Baruch R. Is inversion 16 a prerequisite and id trisomy 22 invariably associated with inversion 16 in AML-M4eo? Cancer Genet Cytogenet. 2000. 121:106.
24.Thirman MJ., Gill HJ., Burnett RC., Mbangkollo D., McCabe NR., Kobayashi H, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993. 329:909–14.
25.Kim HJ., Cho HI., Kim EC., Ko EK., See CJ., Park SY, et al. A study on 289 consecutive Korean patients with acute leukaemias revealed fluorescence in situ hybridization detects the MLL translocation without cytogenetic evidence both initially and during follow-up. Br J Haematol. 2002. 119:930–9.
26.Meyer C., Schneider B., Jakob S., Strehl S., Attarbaschi A., Schnittger S, et al. The MLL recombinome of acute leukemias. Leukemia. 2006. 20:777–84.
Fig. 1.
Karyotype by conventional chromosome analysis (A) and the FISH patterns showing CBFβ break-apart signals (B) shown in case 1.
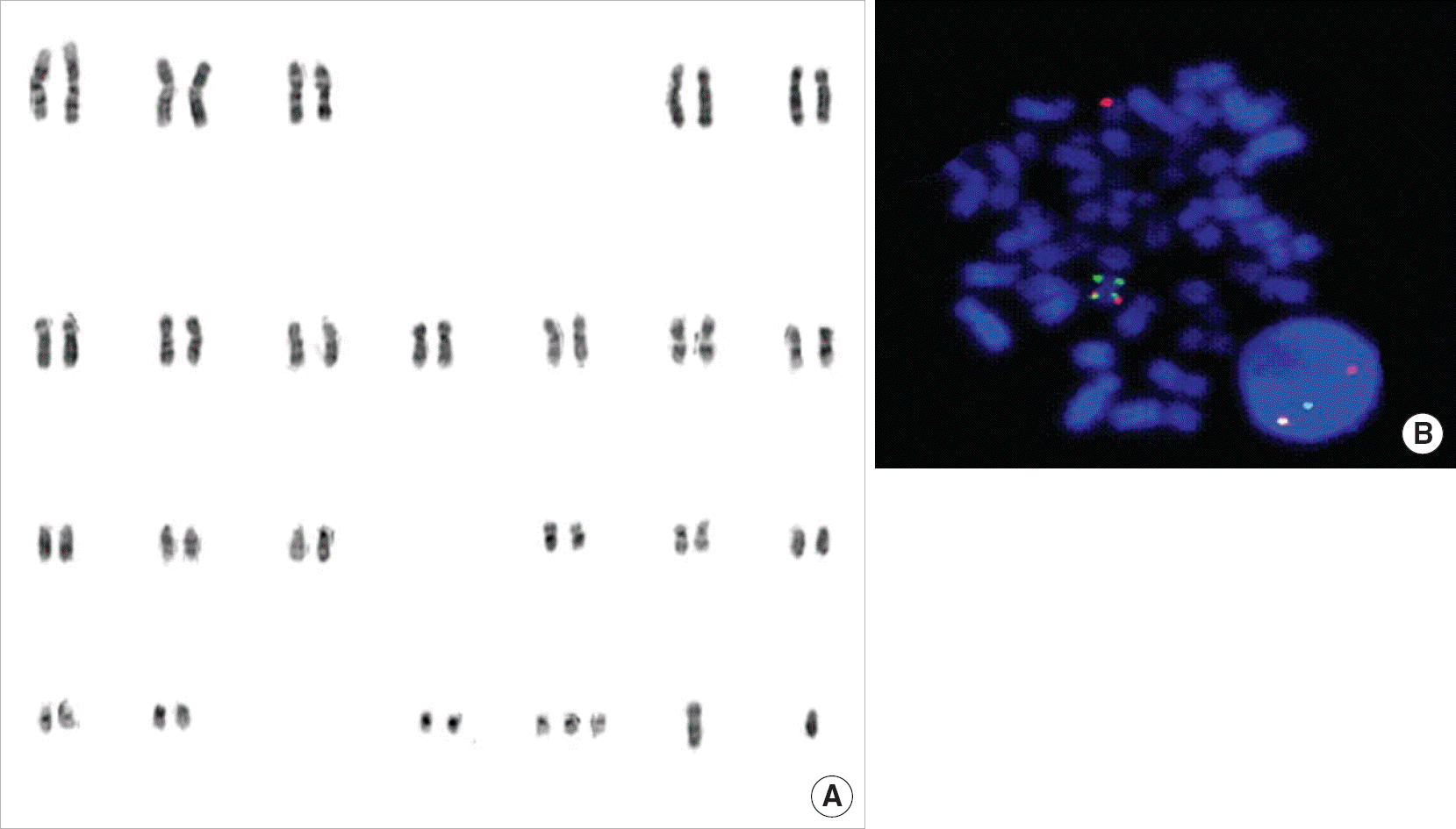
Table 1.
List of the probes used for fluorescence in-situ hybridization analysis in this study
Table 2.
Characteristics of acute leukemia cases where fluorescence in-situ hybridization profile test were applied
| ALL FISH profile test were applied | AML FISH profile test were applied | ||||||
|---|---|---|---|---|---|---|---|
| Parameter/immunophenotype | B-cell type (109)∗ | T-cell type (14)∗ | Biphenotype/mixed lineage (15)∗ | Parameter/classification | AML with recurrent genetic abnormalities (29)∗ | AML, not otherwise specified (65)∗ | Others (AML with MD, etc.) (9)∗ |
| Age (yr) | Age (yr) | ||||||
| Children (age<18) | Children (age<18) | 14 | 10 (M7:4) | 1 | |||
| <1 | 3 | 0 | 0 | Adult | |||
| 1-10 | 61 | 2 | 5 | <55 | 9 | 36 | 6 |
| >10 | 24 | 3 | 2 | >55 | 6 | 19 | 2 |
| Adult | Gender | ||||||
| <65 | 21 | 9 | 8 | Male | 16 | 39 | 7 |
| Gender | Female | 13 | 26 | 2 | |||
| Male | 53 | 9 | 8 | ||||
| Female | 56 | 5 | 7 | ||||
Table 3.
Frequencies of the genetic abnormalities according to the detection methods, conventional cytogenetic analysis (CCA) and FISH profile test
Categories: 1∗, CCA: normal or ‘no mitotic cells’/FISH profile test: normal; 2†, Compatible results of cytogenetic abnormalities between the two detection methods; 3‡, CCA: abnormal / FISH profile test: normal; 4§, Additional cytogenetic abnormalities were detected only by FISH profile test; 5‖, CCA: normal or ‘no mitotic cells’/FISH profile test: abnormal.
Table 4.
Frequencies of the abnormalities identified by ALL FISH profile in 51 acute leukemia cases with normal karyotypes or without cytogenetic results (category 5)
Table 5.
Summary of the results of conventional chromosome analysis and FISH profile test of 4 cases of “AML with recurrent genetic abnormalities” and category 5




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download