Abstract
Background:
Mycobacterium tuberculosis is one of the most clinically significant infectious agents. Especially during mass outbreaks, accurate identification and monitoring are required. The proportion of Beijing family members is very high among infecting strains, and spoligotyping is not suitable for strain typing. Therefore, we studied the homogeneity of isolates using the mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) method and identified its utility for carrying out molecular epidemiologic analysis.
Methods:
Eighty-one clinical M. tuberculosis isolates that had previously been analyzed by spoligotyping were used in this study. We used the 12 standard MIRU loci and further four exact tandem repeat (ETR) loci (ETR-A, -B, -C, and -F). Four strains each of randomly selected Beijing and Beijinglike families were subjected to IS6110- restriction fragment length polymorphism analysis.
Results:
All 81 samples showed amplification products of all VNTR loci, and all of them showed differences in at least one locus. The calculation of the Hunter-Gaston diversity index (HGDI) for MIRU-VNTR gave the value of 0.965. Discriminatory index in the six loci (MIRU-10, −16, −26, −31, −39, and ETR-F) were found to be highly discriminated (HGDI >0.6). Beijing and Beijing-like family isolates were discriminated into different MIRU-VNTR types.
REFERENCES
1.Bloom BR., Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992. 257:1055–64.

2.World Health Organization. Health across the life span. The world health report 1998-Life in the 21st century: A vision for all. Geneva: World Health Organization;1998. p. 61–112.
3.Choi CM., Jeung WK., Kang CI., Kim DH., Kim YK., Heo ST, et al. Clinical characteristics of tuberculosis in North Korean refugees. Tuberc Respir Dis. 2006. 60:285–9. (최창민, 정우경, 강철인, 김도형, 김영근, 허상택 등. 북한이탈주민에서의 결핵의 임상적 고찰. 결핵 및 호흡기질환 2006;60:285-9.).

4.Asgharzadeh M., Kafil HS. Current trends in molecular epidemiology studies of Mycobacterium tuberculosis. Biotechnol Mol Biol Rev. 2007. 2:108–15.
5.Kremer K., van Soolingen D., Frothingham R., Haas WH., Hermans PW., Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999. 37:2607–18.
6.Mazars E., Lesjean S., Banuls AL., Gilbert M., Vincent V., Gicquel B, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA. 2001. 98:1901–6.
7.Kremer K., Au BK., Yip PC., Skuce R., Supply P., Kam KM, et al. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005. 43:314–20.
8.Kamerbeek J., Schouls L., Kolk A., van Agterveld M., van Soolingen D., Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997. 35:907–14.
9.Supply P., Lesjean S., Savine E., Kremer K., van Soolingen D., Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001. 39:3563–71.
10.Frothingham R., Meeker-O'Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998. 144:1189–96.
11.Surikova OV., Voitech DS., Kuzmicheva G., Tatkov SI., Mokrousov IV., Narvskaya OV, et al. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur J Epidemiol. 2005. 20:963–74.
12.Iwamoto T., Yoshida S., Suzuki K., Tomita M., Fujiyama R., Tanaka N, et al. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol Lett. 2007. 270:67–74.
13.Alonso-Rodriguez N., Martinez-Lirola M., Herranz M., Sanchez-Benitez M., Barroso P., Bouza E, et al. Evaluation of the new advanced 15-loci MIRU-VNTR genotyping tool in Mycobacterium tuberculosis molecular epidemiology studies. BMC Microbiol. 2008. 8:34.

14.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rusch-Gerdes S., Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006. 44:4498–510.
15.Song EJ., Jeong HJ., Lee SM., Kim CM., Song ES., Park YK, et al. A DNA chip-based spoligotyping method for the strain identification of Mycobacterium tuberculosis isolates. J Microbiol Methods. 2007. 68:430–3.
16.van Soolingen D., Hermans PW., de Haas PE., Soll DR., van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991. 29:2578–86.
17.Supply P., Mazars E., Lesjean S., Vincent V., Gicquel B., Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000. 36:762–71.
18.Hunter PR., Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988. 26:2465–6.

19.van Embden JD., Cave MD., Crawford JT., Dale JW., Eisenach KD., Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993. 31:406–9.
20.Sola C., Filliol I., Legrand E., Lesjean S., Locht C., Supply P, et al. Geno-typing of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003. 3:125–33.
21.Kremer K., Glynn JR., Lillebaek T., Niemann S., Kurepina NE., Kreiswirth BN, et al. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J Clin Microbiol. 2004. 42:4040–9.
22.Otal I., Martin C., Vincent-Levy-Frebault V., Thierry D., Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J Clin Microbiol. 1991. 29:1252–4.

23.Kremer K., Arnold C., Cataldi A., Gutierrez MC., Haas WH., Panaiotov S, et al. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J Clin Microbiol. 2005. 43:5628–38.
24.Cowan LS., Mosher L., Diem L., Massey JP., Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002. 40:1592–602.
25.Kwara A., Schiro R., Cowan LS., Hyslop NE., Wiser MF., Roahen Harrison S, et al. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003. 41:2683–5.
26.Hawkey PM., Smith EG., Evans JT., Monk P., Bryan G., Mohamed HH, et al. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J Clin Microbiol. 2003. 41:3514–20.
Fig. 1.
MIRU-VNTR electrophoresis patterns of three M. tuberculosis isolates. Lane M, 100-bp molecular marker. Other lanes in each isolate represent the 16 VNTR loci. Order of the 16 VNTR loci is the same as in Table 1 and 2.
Abbreviation: See Table 1.
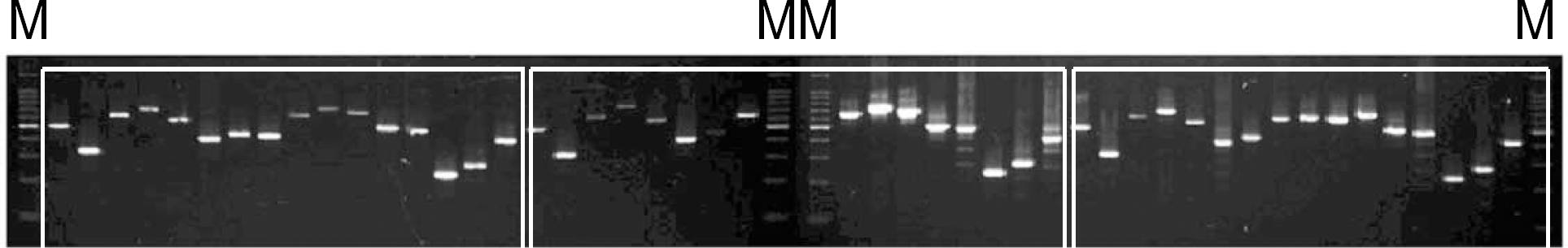
Fig. 2.
Comparison of molecular strain typing patterns of four M. tuberculosis Beijing family isolates showing the diversity of MIRU-VNTR. (A) IS6110 RFLP analysis pattern, (B) Spoligotyping pattern, and (C) MIRU-VNTR pattern. Order of the 16 VNTR loci is the same as in Table 1 and 2. All isolates belonging to the Beijing family defined by Kremer et al. [21] hybridized to the last nine probes in spoligotyping, but differed in at least three loci in MIRU-VNTR analysis.
Abbreviations: RFLP, restriction fragment length polymorphism; MIRU-VNTR, mycobacterial interspersed repetitive units-variable number of tandem repeats.
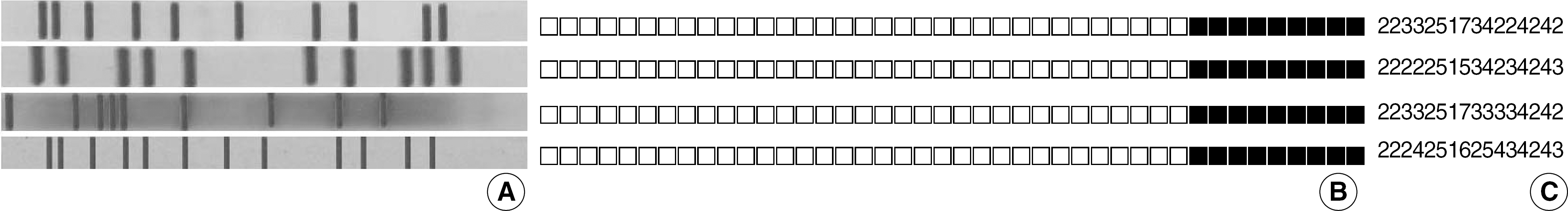
Fig. 3.
Comparison of molecular strain typing patterns of four M. tuberculosis Beijing-like family isolates showing diversity of MIRU-VNTR. (A) IS6110 RFLP analysis pattern, (B) Spoligotyping pattern, and (C) MIRU-VNTR pattern. Order of the 16 VNTR loci is the same as in Table 1 and 2. All isolates belonging to the Beijing-like family defined by Kremer et al. [21] hybridized exclusively with most of the last nine probes in spoligotyping, but differed from each other in at least two loci in MIRU-VNTR analysis.
Abbreviations: See Fig. 2.
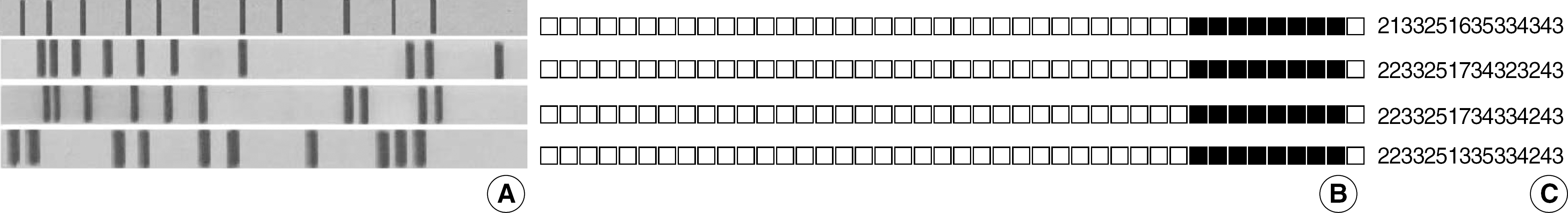
Table 1.
Primer sequences for amplifying MIRU-VNTR loci
Table 2.
Distribution of 81 M. tuberculosis isolates containing variable numbers of each VNTR locus and allelic diversity
| VNTR Loci | N of VNTR repeats | HGDI∗ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||
| MIRU-2 | 18 | 61 | 2 | 0.388 | |||||||
| MIRU-4 | 1 | 3 | 77 | 0.096 | |||||||
| MIRU-10 | 4 | 35 | 34 | 8 | 0.633 | ||||||
| MIRU-16 | 2 | 32 | 35 | 12 | 0.643 | ||||||
| MIRU-20 | 4 | 77 | 0.095 | ||||||||
| MIRU-23 | 6 | 69 | 4 | 1 | 1 | 0.269 | |||||
| MIRU-24 | 78 | 3 | 0.072 | ||||||||
| MIRU-26 | 1 | 12 | 12 | 9 | 12 | 28 | 5 | 2 | 0.808 | ||
| MIRU-27 | 1 | 23 | 50 | 7 | 0.537 | ||||||
| MIRU-31 | 1 | 8 | 17 | 24 | 20 | 11 | 0.789 | ||||
| MIRU-39 | 10 | 34 | 30 | 7 | 0.672 | ||||||
| MIRU-40 | 1 | 25 | 47 | 7 | 1 | 0.567 | |||||
| ETR-A | 2 | 5 | 73 | 1 | 0.185 | ||||||
| ETR-B | 3 | 72 | 6 | 0.206 | |||||||
| ETR-C | 1 | 5 | 74 | 1 | 0.163 | ||||||
| ETR-F | 1 | 10 | 33 | 26 | 10 | 1 | 0.709 | ||||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download