Abstract
Background
Since the human genome project was completed in 2003, there have been numerous reports on cancer and related markers. This study was aimed to develop a system to extract automatically information regarding the relationship between cancer and tumor markers from biomedical literatures.
Methods
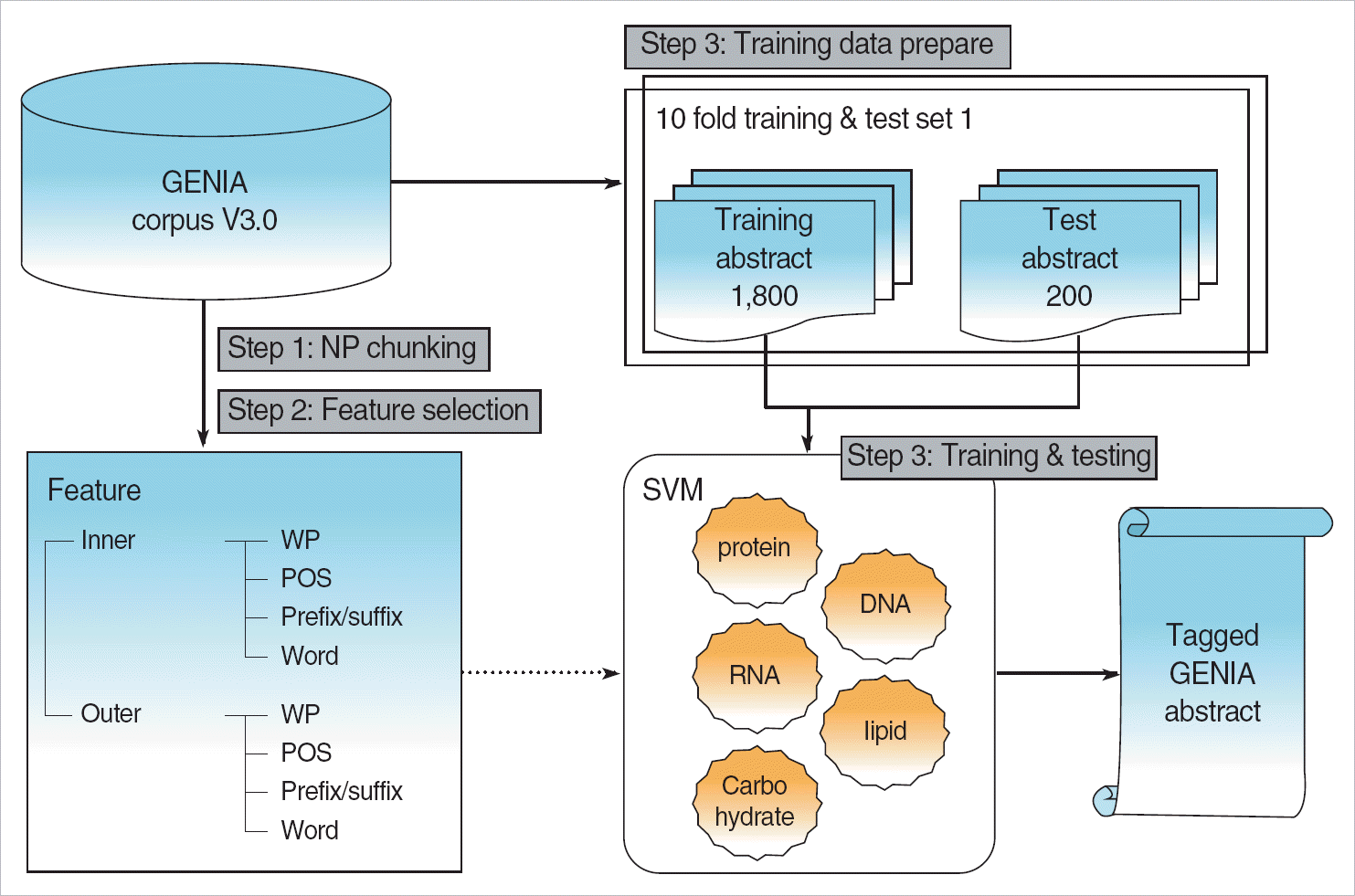
Named entities of tumor markers were recognized by both a dictionary-based method and machine learning technology of the support vector machine. Named entities of cancers were recognized by the MeSH dictionary.
Results
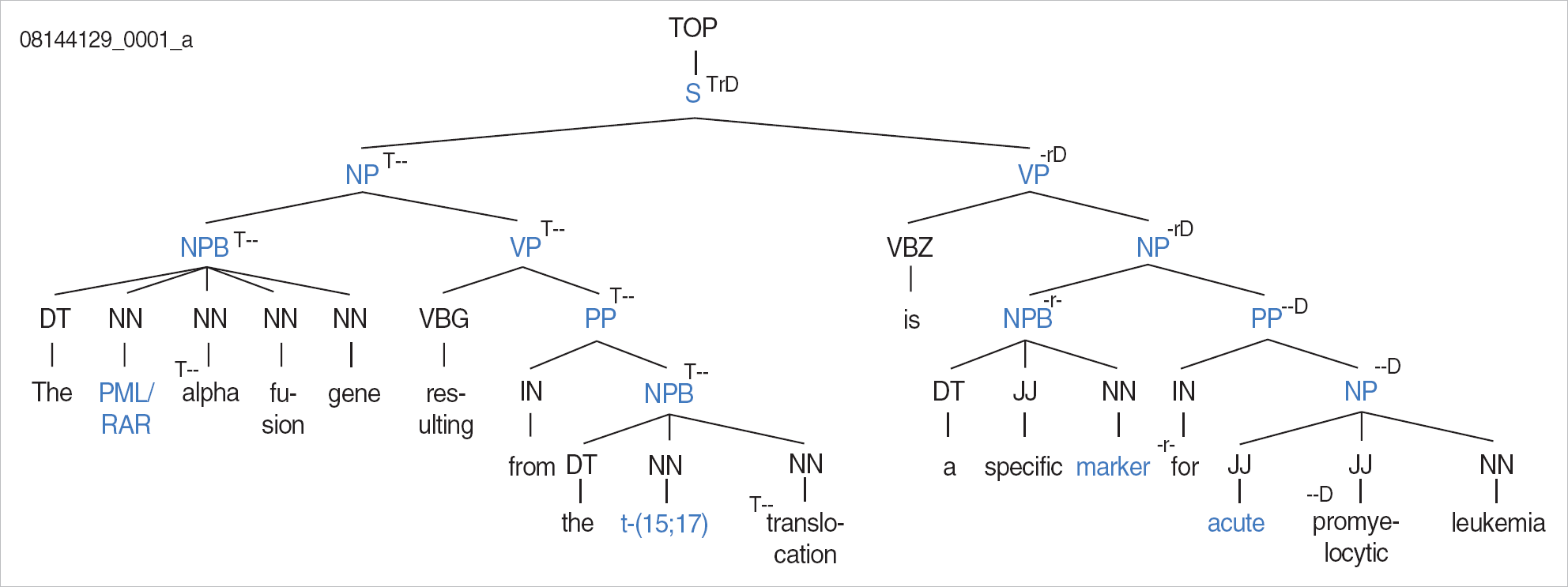
Relational and filtering keywords were selected after annotating 160 abstracts from PubMed. Relational information was extracted only when one of the relational keywords was in an appropriate position along the parse tree of a sentence with both tumor marker and disease entities. The performance of the system developed in this study was evaluated with another set of 77 abstracts. With the relational and filtering keyword used in the system, precision was 94.38% and recall was 66.14%, while without the expert knowledge precision was 49.16% and recall was 69.29%.
Conclusions
We developed a system that can extract relational information between a tumor and its markers by incorporating expert knowledge into the system. The system exploiting expert knowledge would serve as a reference when developing another information extraction system in various medical fields.
REFERENCES
1.Collins FS, Green ED, Guttmacher AE, Guyer MS, US National Human Genome Research Institute. A vision for the future of geno-mics research. Nature. 2003. 422:835–47.
2.Jensen LJ., Saric J., Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet. 2006. 7:119–29.

3.Temkin JM., Gilder MR. Extraction of protein interaction information from unstructured text using a context-free grammar. Bioinformatics. 2003. 19:2046–53.

4.Friedman C., Kra P., Yu H., Krauthammer M., Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001. 17(S):S74–82.

5.Ono T., Hishigaki H., Tanigami A., Takagi T. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics. 2000. 17:155–61.

6.Cristianini N, Shawe-Taylor J, editors. An introduction to support vector machines and other kernel based learning methods. 1st ed.Cambridge: Cambridge University Press;2000.
7.McNaught J., Black WJ. Information extraction. Ananiadou S, McNaught J, editors. Text Mining for biology and biomedicine. 1st ed.Norwood: Artech House;2006. p. 143–77.
8.Kim JD., Ohta T., Tateisi Y., Tsujii J. GENIA corpus- semantically annotated corpus for bio-textmining. Bioinformatics. 2003. 19(S):i180–2.
9.Collins M. Head-Driven Statistical Models for Natural Language Parsing [Dissertation]. Philadelphia (PA): Pennsylvania Univ.;1995.
10.Tanabe L., Wilbur WJ. Tagging gene and protein names in biomedical text. Bioinformatics. 2002. 18:1124–32.

11.Kazama J., Makino T., Ohta Y., TsujiiJ J. Tunning support vector machines for biomedical named entity recognition. In: Association for Computational Linguistics, ed. ACL 2002 Workshop. Proceedings of the ACL 2002 Workshop on Natural Language Processing in the Biomedical Domain; 2002 July 11; Philadelphia, PA, USA;. 2002. 1–8.
12.Zhou G., Zhang J., Su J., Shen D., Tan C. Recognizing names in biomedical texts: a machine learning approach. Bioinformatics. 2004. 20:1178–90.

13.Proux D., Rechenmann F., Julliard L., Pillet V V., Jacq B. Detecting Gene Symbols and Names in Biological Texts: A First Step toward Pertinent Information Extraction. Genome Inform Ser Workshop Genome Inform. 1998. 9:72–80.
14.Chae JM, Jung SY, Oh HB. (Eds.).Tumor marker information extraction system. http://medtextmining.net/. (Updated on Aug 2006).
15.Ananiadou S., McNaught J. Introduction. Ananiadou S, McNaught J, editors. Text mining for biology and biomedicine. 1st ed.Norwood: Artech House;2006. p. 1–12.
16.Lee KJ., Hwang YS., Kim S., Rim HC. Biomedical named entity recognition using two-phase model based on SVMs. J Biomed Inform. 2004. 37:436–47.

17.Park JC., Kim JJ. Named entity recognition. Ananiadou S, McNaught J, editors. Text mining for biology and biomedicine. 1st ed.Norwood: Artech House;2006. p. 121–42.
18.Ananiadou S., Nenadic G. Automatic terminology management in biomedicine. Ananiadou S, McNaught J, editors. Text mining for biology and biomedicine. 1st ed.Norwood: Artech House, Inc.;2006. p. 67–98.
19.Bodenreider O. Lexical, terminological, and ontological resources for bilogical text mining. Ananiadou S, McNaught J, editors. Text mining for biology and biomedicine. 1st ed.Norwood: Artech House;2006. p. 43–67.
20.Hatzivassiloglou V., Duboue PA., Rzhetsky A. Disambiguating proteins, genes, and RNA in text: a machine learning approach. Bioinformatics. 2001. 17(S):S97–106.

21.Krauthammer M., Rzhetsky A., Morozov P., Friedman C. Using BLAST for identifying gene and protein names in journal articles. Gene. 2000. 259:245–52.

22.Marsh SG., Albert ED., Bodmer WF., Bontrop RE., Dupont B., Erlich HA, et al. Nomenclature for factors of the HLA system, 2004. Tissue Antigens. 2005. 65:301–69.

23.Horn F., Lau AL., Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004. 20:557–68.

24.Novichkova S., Egorov S., Daraselia N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics. 2003. 19:1699–706.

25.Shin HR., Won YJ., Jung KW., Park JG., Ahn YO. Cancer Registration and Statistics in Korea. J Korean Assoc Cancer Prev. 2004. 9:49–55. (신해림, 원영주, 정규원, 박재갑, 안윤옥. 우리나라 암등록사업과암통계. 대한암예방학회지 2004;9: 49-55.).
Fig. 1.
System overview for the relational information extraction. (A) Named entity recognition module. (B) Information extraction module.
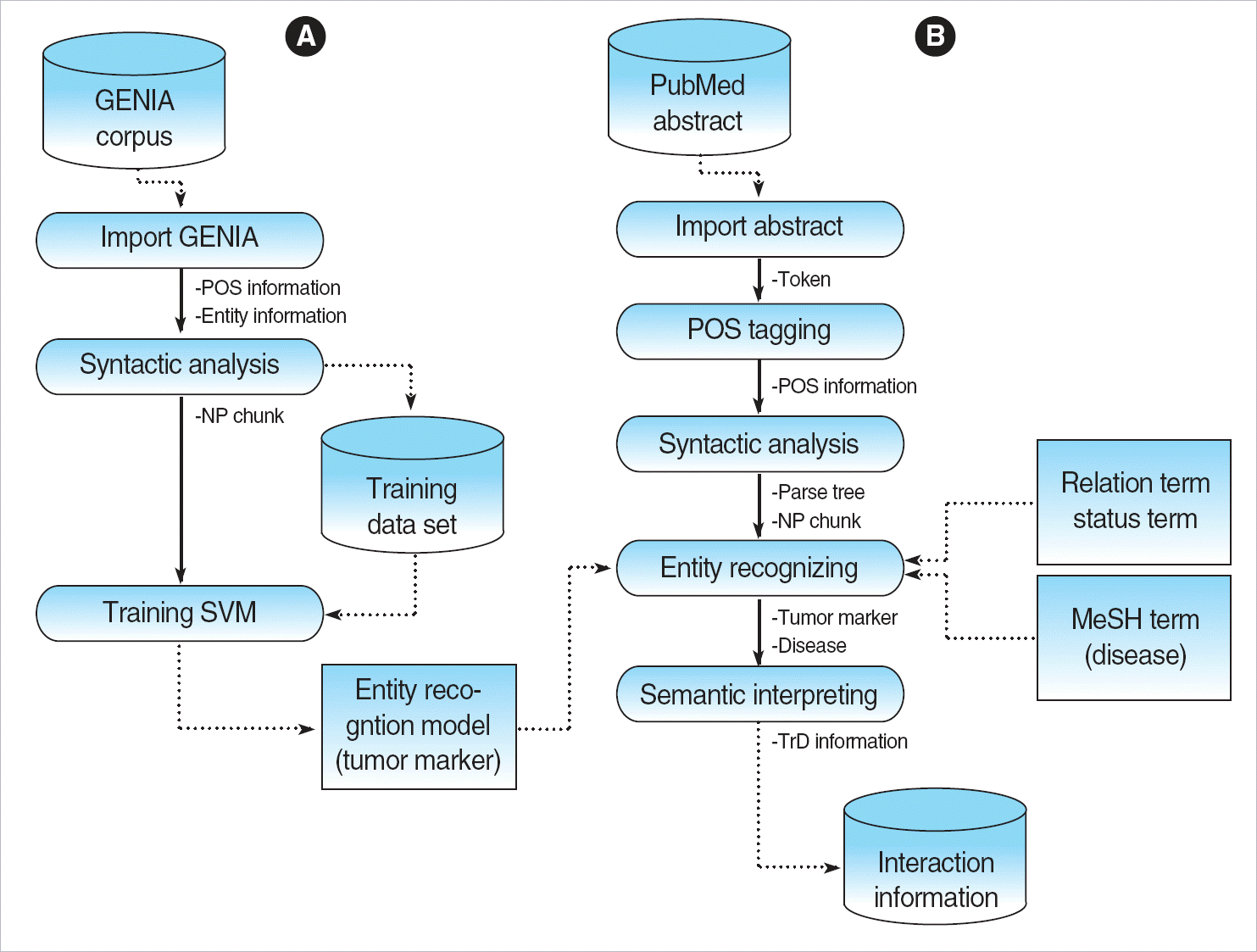
Table 1.
Recognizing methods for named biological entities
NCI Thesaurus (http://nciterms.nci.nih.gov/NCIBrowser/Dictionary.do).
Medical Subject Heading (MeSH) (http://www.nlm.nih.gov/mesh/).
Table 2.
An example of relation and filtering keywords for extracting information relating between tumors and tumor markers




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download