Abstract
Background
Exon deletions of Duchenne muscular dystrophy (DMD) gene account for most of the alterations found in DMD and Becker muscular dystrophy (BMD). This study was to evaluate the usefulness of dual priming oligonucleotide multiplex PCR (DPO PCR) in detection of exon deletions of DMD gene.
Methods
Thirty-seven DMD or BMD patients who had known exon deletions detected by conventional multiplex PCR (conventional PCR) and nine control subjects were enrolled in this study. When a discrepancy was shown between the results of conventional PCR and DPO PCR, the multiplex ligation-dependent probe amplification (MLPA) technique was performed as a confirmation test.
Results
The same deletions previously identified by conventional PCR in 32 out of 37 subjects were also detected by DPO PCR. For the five subjects (13.5%) showing discrepant results between the conventional PCR and DPO PCR, MLPA was performed and its results were found to correlate better with those of DPO PCR. The discrepancies were due to false positive or false negative results of the conventional PCR.
Conclusions
DPO PCR shows a high agreement of results with the conventional PCR and is considered an adequate method to be used as a primary genetic test for the diagnosis of DMD. Because of an improved accuracy, especially for determining the boundaries of DMD gene deletions, DPO PCR can be very useful as a supplement to the conventional PCR.
REFERENCES
1.Emery AE. Population frequencies of inherited neuromuscular diseases–a world survey. Neuromuscul Disord. 1991. 1:19–29.

2.Abbs S., Yau SC., Clark S., Mathew CG., Bobrow M. A convenient multiplex PCR system for the detection of dystrophin gene deletions: a comparative analysis with cDNA hybridisation shows mistypings by both methods. J Med Genet. 1991. 28:304–11.

3.Den Dunnen JT., Grootscholten PM., Dauwerse JG., Walker AP., Monaco AP., Butler R, et al. Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination. Hum Mol Genet. 1992. 1:19–28.
4.Nobile C., Marchi J. A refined restriction map of YAC clones spanning the entire human dystrophin gene. Mamm Genome. 1994. 5:566–71.

5.Ervasti JM., Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991. 66:1121–31.

6.Ervasti JM., Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993. 122:809–23.

7.Choi JR., Song KS., Park SJ. Genetic polymorphism analysis for the detection of Duchenne muscular dystrohpy carriers. Korean J Clin Pathol. 2000. 20:236–41. (최종락, 송경순, 박숙자. 듀센형 근이영양증 보인자진단을위한유전자다형분석. 대한임상병리학회지 2000;20: 236-41.).
8.van Essen AJ., Kneppers AL., van der Hout AH., Scheffer H., Ginjaar IB., ten Kate LP, et al. The clinical and molecular genetic approach to Duchenne and Becker muscular dystrophy: an updated protocol. J Med Genet. 1997. 34:805–12.

9.Den Dunnen JT., Grootscholten PM., Bakker E., Blonden LA., Ginjaar HB., Wapenaar MC, et al. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989. 45:835–47.
10.Koenig M., Hoffman EP., Bertelson CJ., Monaco AP., Feener C., Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987. 50:509–17.

11.AH Koenig M., Boyce FM., Kunkel LM. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990. 86:45–8.

12.Park SY., Koh KN., Lim BC., Kang HS., Lee KY., Hwang H, et al. Molecular genetic analysis of dystrophin gene in Duchenne/Becker muscular dystrophy. J Korean Child Neurol Soc. 2004. 12:50–8. (박수연, 고경남, 임병찬, 강호석, 이경연, 황희 등. Duchenne/Becker 근이영양증에서의 Dystrophin 유전자분석. 대한소아신경학회지 2004;12: 50-8.).
13.Chun JY., Kim KJ., Hwang IT., Kim YJ., Lee DH., Lee IK, et al. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007. 35:e40.

14.Lai KK., Lo IF., Tong TM., Cheng LY., Lam ST. Detecting exon deletions and duplications of the DMD gene using Multiplex Ligation-dependent Probe Amplification (MLPA). Clin Biochem. 2006. 39:367–72.

15.Chamberlain JS., Gibbs RA., Ranier JE., Caskey CT. Multiplex PCR for the diagnosis of Duchenne muscular dystrophy. Innis MA, Gelfand DH, editors. PCR Protocols: A Guide to Methods and Applications. 1st ed.San Diego: Academic Press;1990. p. 272–81.

16.Fujimura FK., Northrup H., Beaudet AL., O'Brien WE. Genotyping errors with the polymerase chain reaction. N Engl J Med. 1990. 322:61.

17.Bergstrom DE., Zhang P., Johnson WT. Comparison of the base pairing properties of a series of nitroazole nucleobase analogs in the oligodeoxyribonucleotide sequence 5′-d(CGCXAATTYGCG)-3′. Nucleic Acids Res. 1997. 25:1935–42.
Fig. 1.
Multiplex ligation-dependent probe amplification (MLPA), conventional-multiplex PCR, and dual priming oligonucleotide (DPO) multiplex PCR of case 34. (A) Deletion of exons 48 and 49 of the DMD gene (arrows) in MLPA. (B) Deletion of exon 49 in lane 2 and presence of exon 48 band in lane 1 of conventional-multiplex PCR. (C) Deletion of exon 48 in group 3 DPO multiplex PCR. M, the 100-bp-size marker.
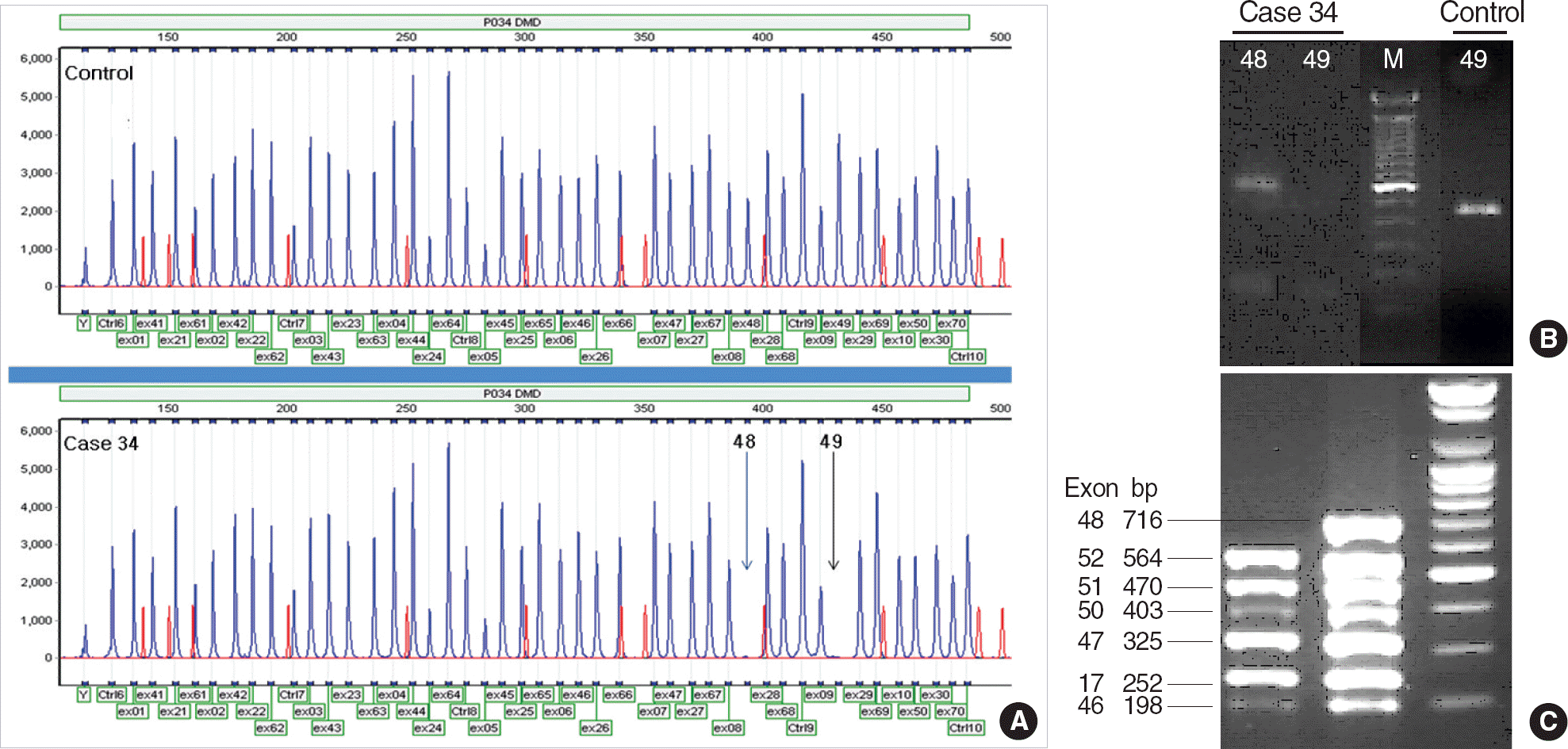
Table 1.
Concordant results between conventional multiplex PCR and DPO multiplex PCR
Table 2.
Results of MLPA performed on the five cases that produced discrepant results between conventional-multiplex PCR and DPO multiplex PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download