Abstract
Background
In previous studies, most hepatitis A virus (HAV) isolates had been genotype IA in Korea. Recently, a small number of different genotypes were reported with an upsurge of acute hepatitis by HAV. We investigated the distribution of HAV genotypes.
Methods
RNA was extracted from anti-HAV IgM positive sera which were collected from March 2007 to February 2008 at a tertiary care hospital in Northeastern Seoul, Korea. Nested reverse transcription (RT)-PCR and direct sequencing for VP1/P2A region of the HAV were performed.
Results
A total of 699 cases with suspected acute hepatitis were tested for anti-HAV IgM, and positive results were obtained in 56 sera (8.0%), which were collected 2 to 15 days (median, 7 days) after the onset of symptoms. Of the 56 seropositive samples, 52 (92.9%) were positive for HAV RNA, among which 28 isolates (53.8%) belonged to genotype IA and the remaining 24 (46.2%) belonged to genotype IIIA. Both IA and IIIA genotypes were isolated from 6-7 neighboring administrative districts throughout the year without geographic or seasonal restrictions.
Go to : 
REFERENCES
1.Park CH., Cho YK., Park JH., Jun JS., Park ES., Seo JH, et al. Changes in the age-specific prevalence of hepatitis A virus antibodies: a 10-year cohort study in Jinju, South Korea. Clin Infect Dis. 2006. 42:1148–50.

2.Kang CI., Choi CM., Park TS., Lee DJ., Oh MD., Choi KW. Incidence and seroprevalence of hepatitis A virus infections among young Korean soldiers. J Korean Med Sci. 2007. 22:546–8.

3.Barzaga BN. Hepatitis A shifting epidemiology in South-East Asia and China. Vaccine. 2000. 18(S):S61–4.

4.Korea Center for Disease Control and Prevention. Changing patterns of hepatitis A virus infection in Korea. Public Health Weekly Report. 2008. 1:169–72. (질병관리본부. 최근 우리나라 A형간염 발생의변화양상. 주간건강과질병 2008;1: 169-72).
5.Nainan OV., Xia G., Vaughan G., Margolis HS. Diagnosis of hepatitis a virus infection: a molecular approach. Clin Microbiol Rev. 2006. 19:63–79.

6.Korea Center for Disease Control and Prevention. An outbreak of hepatitis A in Gongju-si. communicable disease monthly report. 2004. 15:205–12. (질병관리본부. 공주시A형간염 유행. 감염병발생정보 2004;15: 205-12.).
7.Park JY., Lee JB., Jeong SY., Lee SH., Lee MA., Choi HJ. Molecular characterization of an acute hepatitis A outbreak among healthcare workers at a Korean hospital. J Hosp Infect. 2007. 67:175–81.

8.Jee YM., Go U., Cheon D., Kang Y., Yoon JD., Lee SW, et al. Detection of hepatitis A virus from clotting factors implicated as a source of HAV infection among haemophilia patients in Korea. Epidemiol Infect. 2006. 134:87–93.

9.Bower WA., Nainan OV., Han X., Margolis HS. Duration of viremia in hepatitis A virus infection. J Infect Dis. 2000. 182:12–7.

10.Kim JS., Kim SH. Molecular epidemiology of an outbreak of hepatitis A in Korea. Korean J Clin Pathol. 2001. 21:114–8. (김재석및김상훈. 집단발병한 A형 간염 바이러스의 분자생물학적 역학연구. 대한임상병리학회지 2001;21: 114-8.).
11.Byun KS., Kim JH., Song KJ., Baek LJ., Song JW., Park SH, et al. Molecular epidemiology of hepatitis A virus in Korea. J Gastroenterol Hepatol. 2001. 16:519–24.
12.Korea Center for Disease Control and Prevention. An outbreak of hepatitis A in Ansan-Si communicable disease monthly report. 2006. 17:12. (질병관리본부. 경기도안산시A형간염 집단 발생 사례. 감염병발생정보 2006;17: 12.).
13.Yun H., Kim S., Lee H., Byun KS., Kwon SY., Yim HJ, et al. Genetic analysis of HAV strains isolated from patients with acute hepatitis in Korea, 2005-2006. J Med Virol. 2008. 80:777–84.

14.Takahashi H., Yotsuyanagi H., Yasuda K., Koibuchi T., Suzuki M., Kato T, et al. Molecular epidemiology of hepatitis A virus in metropolitan areas in Japan. J Gastroenterol. 2006. 41:981–6.

15.Endo K., Inoue J., Takahashi M., Mitsui T., Masuko K., Akahane Y, et al. Analysis of the full-length genome of a subgenotype IIIB hepatitis A virus isolate: primers for broadly reactive PCR and genotypic analysis. J Med Virol. 2007. 79:8–17.

16.Lemon SM., Murphy PC., Shields PA., Ping LH., Feinstone SM., Cromeans T, et al. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991. 65:2056–65.

17.Stene-Johansen K., Jonassen TO., Skaug K. Characterization and genetic variability of Hepatitis A virus genotype IIIA. J Gen Virol. 2005. 86:2739–45.

18.Fujiwara K., Yokosuka O., Fukai K., Imazeki F., Saisho H., Omata M. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J Hepatol. 2001. 35:112–9.

19.Pina S., Buti M., Jardi R., Clemente-Casares P., Jofre J., Girones R. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J Gen Virol. 2001. 82:2955–63.

20.Heo NY., Lim YS., Kang JM., Oh SI., Park CS., Jung SW, et al. Clinical features of fulminant hepatic failure in a tertiary hospital with a liver transplant center in Korea. Korean J Hepatol. 2006. 12:82–92. (허내윤, 임영석, 강정민, 오세일, 박찬선, 정석원 등. 간이식센터가 있는 3차의료기관에 내원한 전격 간부전 환자의 임상적 특징. 대한간학회지 2006;12: 82-92.).
21.Ngui SL., Granerod J., Jewes LA., Crowcroft NS., Teo CG. Outbreaks of hepatitis A in England and Wales associated with two co-circulating hepatitis A virus strains. J Med Virol. 2008. 80:1181–8.

22.Cristina J., Costa-Mattioli M. Genetic variability and molecular evolution of hepatitis A virus. Virus Res. 2007. 127:151–7.

23.Taylor MB. Molecular epidemiology of South African strains of hepatitis A virus: 1982-1996. J Med Virol. 1997. 51:273–9.

24.Bruisten SM., van Steenbergen JE., Pijl AS., Niesters HG., van Doornum GJ., Coutinho RA. Molecular epidemiology of hepatitis A virus in Amsterdam, the Netherlands. J Med Virol. 2001. 63:88–95.

25.Massoudi MS., Bell BP., Paredes V., Insko J., Evans K., Shapiro CN. An outbreak of hepatitis A associated with an infected foodhandler. Public Health Rep. 1999. 114:157–64.

Go to : 
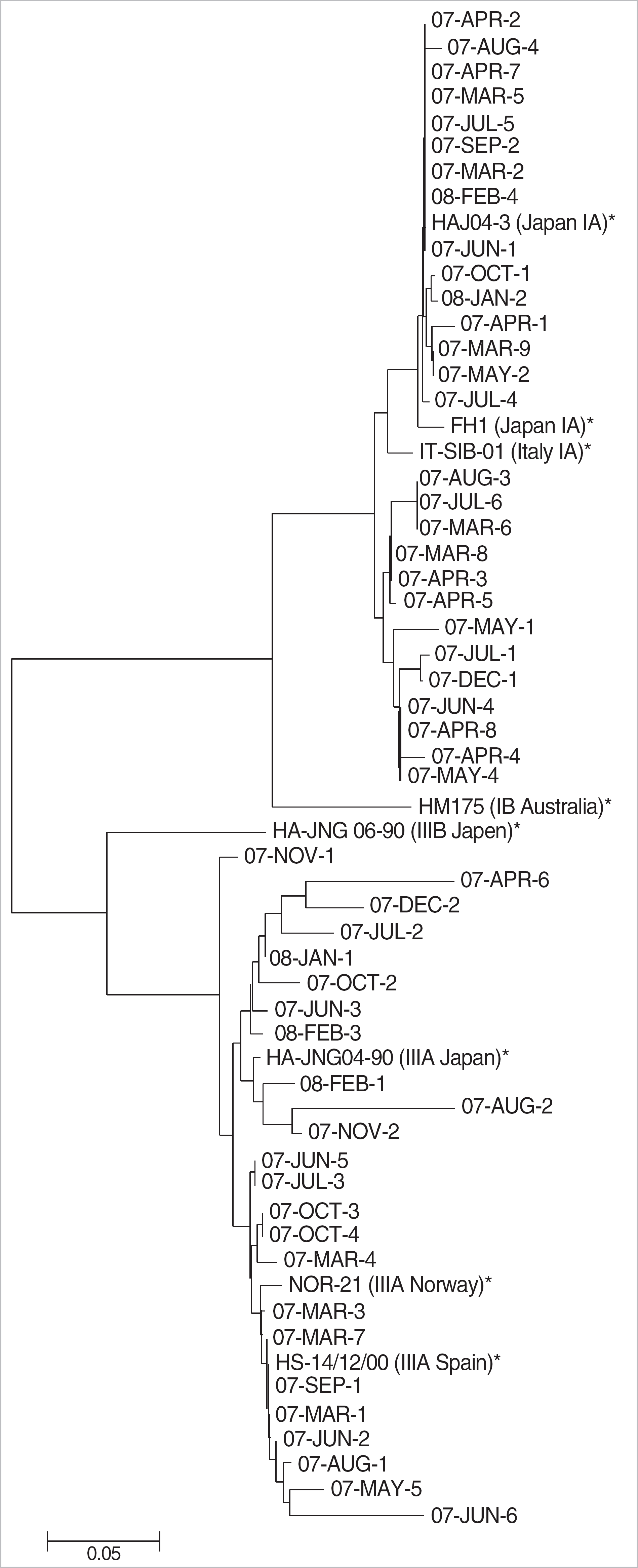 | Fig. 1.A phylogenetic tree constructed for the nucleotide sequences of VP1/P2A region of hepatitis A virus genome. The tree includes 52 strains of Korean patients and reference strains. ∗shows the reference strains acquired abroad; HAJ04-3, FH1, IT-SIB-01 in genotype IA, HM175 in genotype IB, NOR-21, HA-JNG04-90, HS-14/12/00 in genotype IIIA, and HA-JNG06-90 in genotype IIIB. |
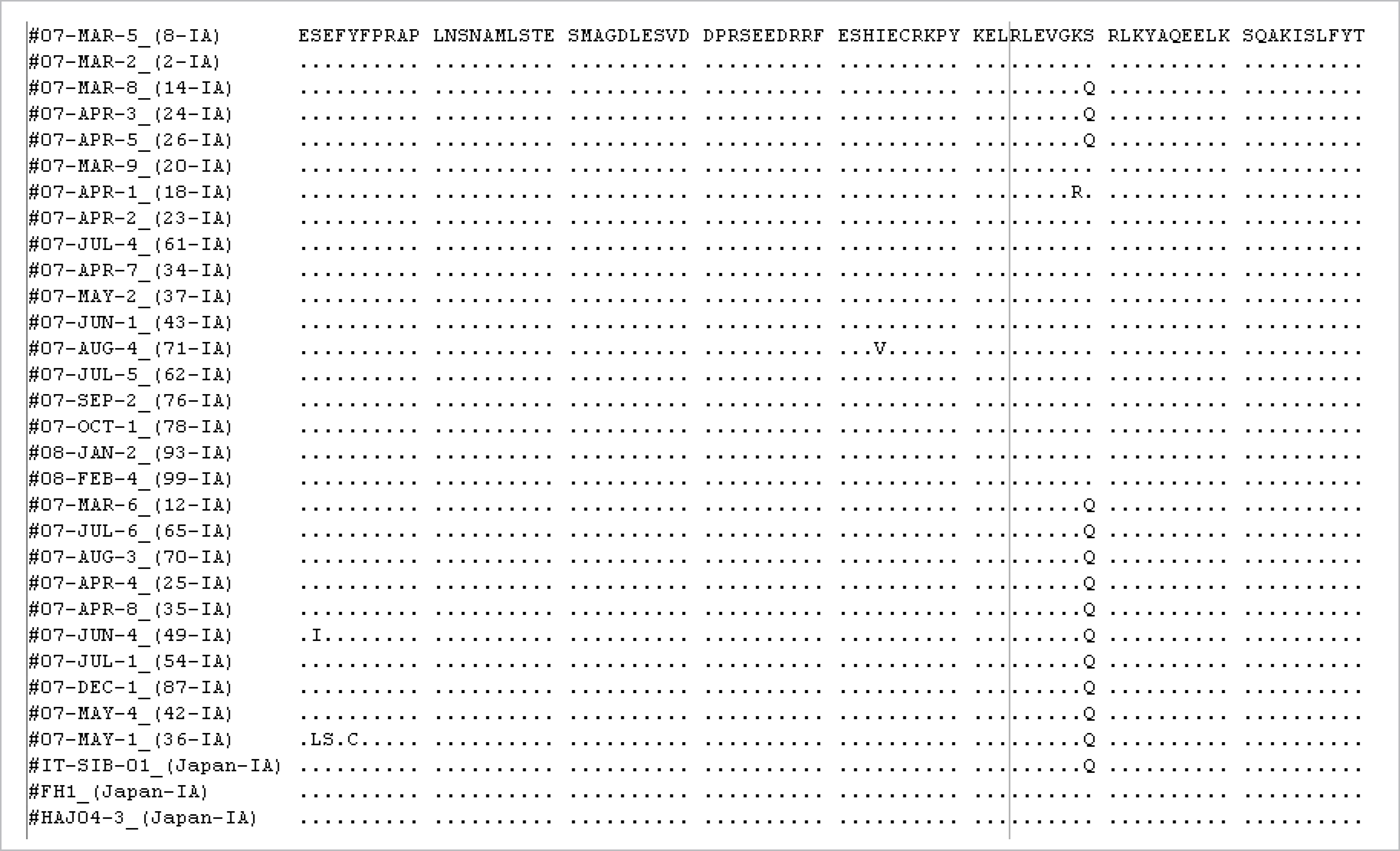 | Fig. 2.Comparison of predicted amino acid sequences of genotype IA isolates. The consensus amino acid sequence for the predominant genotype IA is shown on the top line. Dots indicate conserved amino acids; differences are shown by the appropriate single letter amino acid code. |
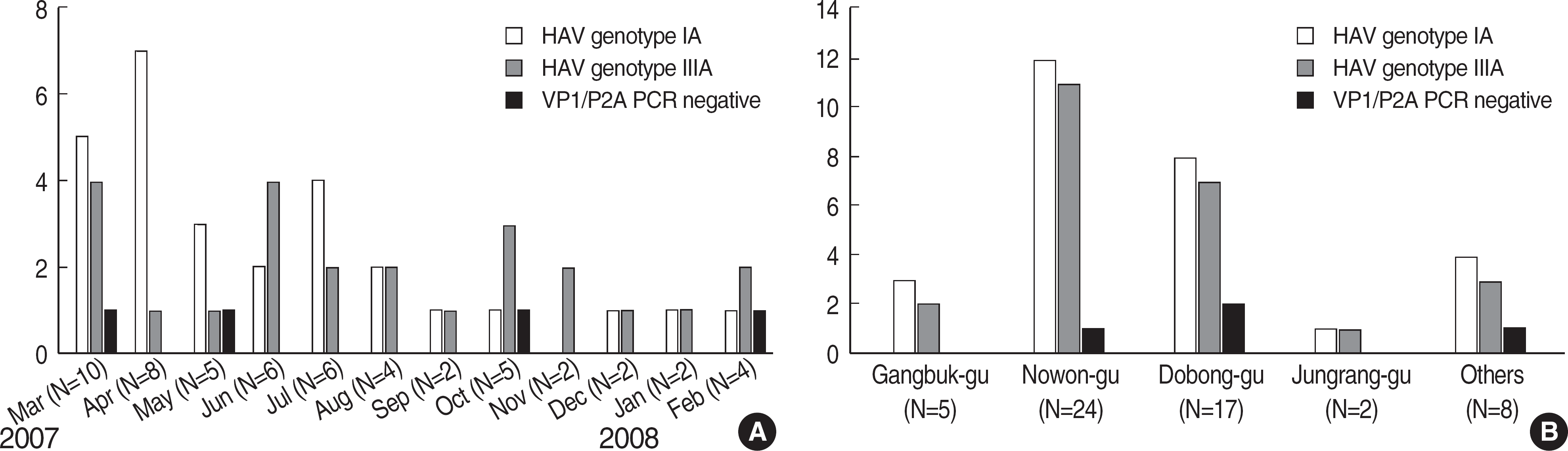 | Fig. 3.Monthly (A) and geographical (B) distribution of 56 patients with hepatitis A. Genotype IA was isolated in 28 patients and genotype IIIA in 24 patients. The remaining 4 patients were negative for VP1/P2A PCR: (A) Monthly distribution from March 2007 to February 2008, (B) Geographic distribution of the patients’ residential areas. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download