Abstract
Background
Coagulase is produced by all strains of Staphylococcus aureus. The 3′ coding region of the coagulase (coa) gene contains varying numbers of 81 bp tandem repeats. S. aureus produces a variety of extracellular protein toxins. Here, we typed S. aureus strains isolated from blood by coa gene restriction fragment length polymorphism (RFLP) patterns and toxin gene profiles.
Methods
A total of 120 strains of S. aureus were isolated from blood cultures during 2003-2006 at Kangdong Sacred Heart Hospital. The isolates were typed by PCR RFLP analysis of the coa gene and by multiplex PCR for detection of genes encoding enterotoxins (sea, seb, sec, sed, and see), toxic shock syndrome toxin-1 (tst), exfoliative toxins (eta and etb), mecA and femA.
Results
All the S. aureus strains were classified into 16 types on the basis of coa gene RFLP and could be further differentiated into 34 types according to the combined patterns of coa gene RFLP and toxin gene profiles. Of 85 methicillin-resistant S. aureus (MRSA) strains, 43 (50.6%) and 36 (42.4%) belonged to the RFLP pattern L5 and pattern L1, respectively. MRSA strains belonging to pattern L5 frequently carried tst (93.0%) or sec gene (81.4%), and strains belonging to pattern L1 frequently carried sea (88.9%) or see gene (44.4%). The rate of the pattern L5 in MRSA strains increased over the past few years and was higher in intensive care unit than in other wards.
Conclusions
We typed S. aureus strains isolated from blood on the basis of coa gene RFLP and toxin genes. The strains belonging to coa gene RFLP pattern L5 and L1 appeared to be the major types of MRSA isolasted from bacteremia and revealed specific toxin gene profiles according to the coa gene RFLP patterns.
Go to : 
REFERENCES
1.Kim JS., Kim HS., Song W., Cho HC., Lee KM., Kim EC. Antimicrobial resistance profiles of Staphylococcus aureus isolated in 13 Korean hospitals. Korean J Lab Med. 2004. 24:223–9. (김재석, 김한성, 송원근, 조현찬, 이규만, 김의종. 국내 13개 의료기관에서 수집된 Staphylococcus aureus의 항균제 감수성 양상. 대한진단검사의학회지 2004;24: 223-9.).
2.Bannerman TL., Peacock SJ. Staphylococcus, Micrococcus and other catalase-positive cocci. Murray PR, Baron EJ, editors. , eds.Manual of Clinical Microbiology. 9th ed.Washington DC: ASM Press;2007. p. 392–402.
3.Kloos WE., Schleifer KH. Genus Staphylococcus. In:. Sneath PHA, Mair NS, editors. Bergey's manual of systematic bacteriology. 1st ed.Baltimore: Williams and Wilkins;1986. p. 1013–35.
4.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998. 339:520–32.
5.Kaida S., Miyata T., Yoshizawa Y., Igarashi H., Iwanaga S. Nucleotide and deduced amino acid sequences of staphylocoagulase gene from Staphylococcus aureus strain 213. Nucleic Acids Res. 1989. 17:8871.
6.Kawabata S., Morita T., Miyata T., Iwanaga S., Igarashi H. Isolation and characterization of staphylocoagulase chymotryptic fragment. Localization of the procoagulant- and prothrombin-binding domain of this protein. J Biol Chem. 1986. 261:1427–33.

7.Goh SH., Byrne SK., Zhang JL., Chow AW. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992. 30:1642–5.
8.Chiou CS., Wei HL., Yang LC. Comparison of pulsed-field gel electrophoresis and coagulase gene restriction profile analysis techniques in the molecular typing of Staphylococcus aureus. J Clin Microbiol. 2000. 38:2186–90.
9.Hookey JV., Richardson JF., Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998. 36:1083–9.
10.Ishino K., Tsuchizaki N., Ishikawa J., Hotta K. Usefulness of PCR-restriction fragment length polymorphism typing of the coagulase gene to discriminate arbekacin-resistant methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007. 45:607–9.
11.Lawrence C., Cosseron M., Mimoz O., Brun-Buisson C., Costa Y., Samii K, et al. Use of the coagulase gene typing method for detection of carriers of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996. 37:687–96.
12.Dinges MM., Orwin PM., Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000. 13:16–34.
13.Thomas D., Chou S., Dauwalder O., Lina G. Diversity in Staphylococcus aureus enterotoxins. Chem Immunol Allergy. 2007. 93:24–41.
14.Ladhani S., Joannou CL., Lochrie DP., Evans RW., Poston SM. Clinical, microbial, and biochemical aspects of the exfoliative toxins causing staphylococcal scalded-skin syndrome. Clin Microbiol Rev. 1999. 12:224–42.

15.Johnson WM., Tyler SD., Ewan EP., Ashton FE., Pollard DR., Rozee KR. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991. 29:426–30.
16.Becker K., Roth R., Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J Clin Microbiol. 1998. 36:2548–53.
17.Mehrotra M., Wang G., Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000. 38:1032–5.
18.Schwarzkopf A., Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994. 32:2407–12.
19.Shopsin B., Gomez M., Waddington M., Riehman M., Kreiswirth BN. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strain. J Clin Microbiol. 2000. 38:3453–6.
20.Tenover FC., Arbeit R., Archer G., Biddle J., Byrne S., Goering R, et al. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994. 32:407–15.
21.Carles-Nurit MJ., Christophle B., Broche S., Gouby A., Bouziges N., Ramuz M. DNA polymorphisms in methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1992. 30:2092–6.
22.Kim JS., Song W., Kim HS., Cho HC., Lee KM., Choi MS, et al. Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn Microbiol Infect Dis. 2006. 56:289–95.
23.Schmitz FJ., MacKenzie CR., Geisel R., Wagner S., Idel H., Verhoef J, et al. Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin sensitive Staphylococcus aureus strains. Eur J Epidemiol. 1997. 13:699–708.
24.Lehn N., Schaller E., Wagner H., Kronke M. Frequency of toxic shock syndrome toxin- and enterotoxin-producing clinical isolates of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1995. 14:43–6.
25.Nishi J., Yoshinaga M., Miyanohara H., Kawahara M., Kawabata M., Motoya T, et al. An epidemiologic survey of methicillin-resistant Staphylococcus aureus by combined use of mec-HVR genotyping and toxin genotyping in a university hospital in Japan. Infect Control Hosp Epidemiol. 2002. 23:506–10.
Go to : 
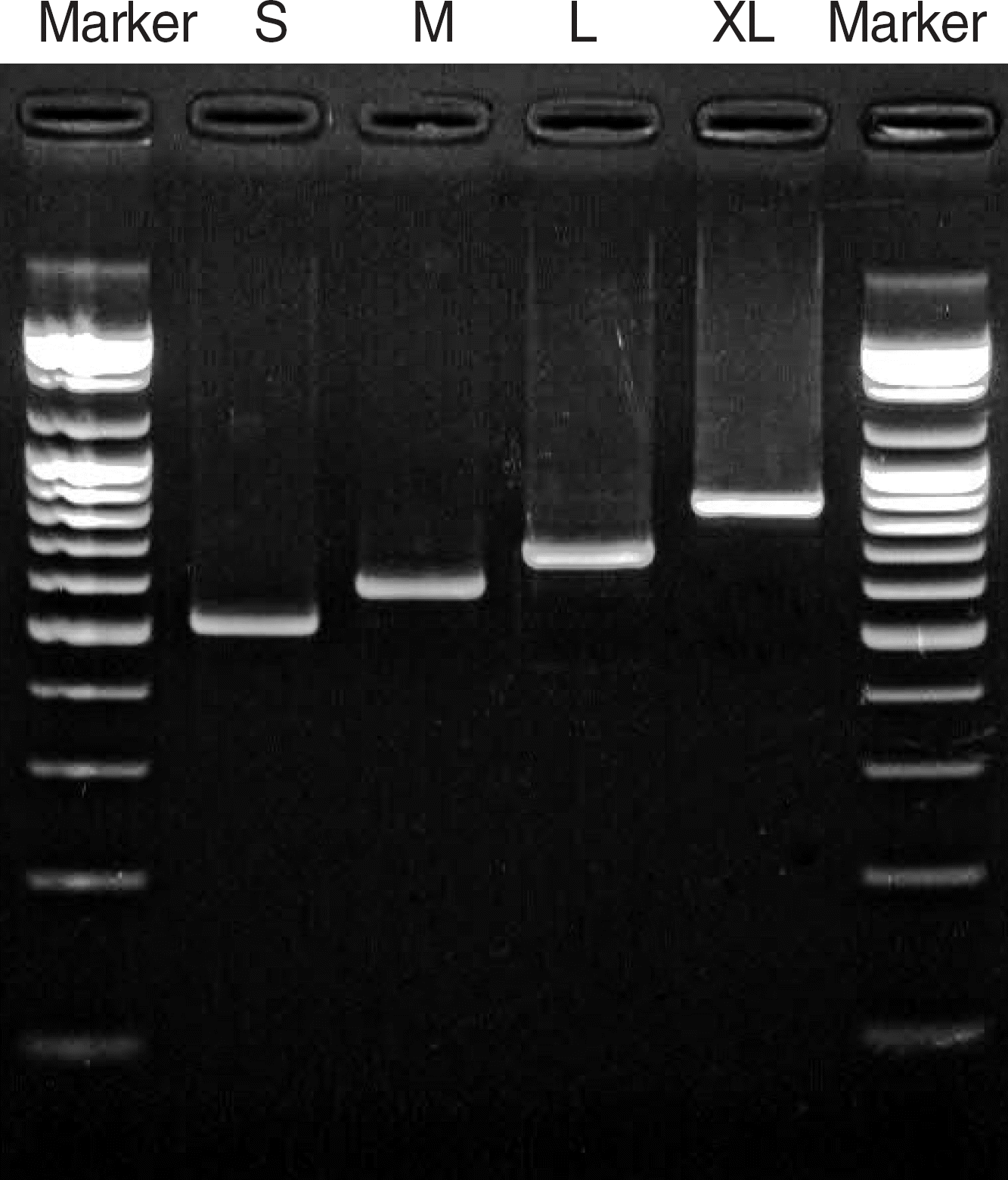 | Fig. 1.Agarose gel electrophoresis of PCR-amplified coa genes from S. aureus strains. RCR amplification with primers COA-1 and COA-2 [9] resulted in single fragment bands of four different sizes: S, M, L, and XL. Marker Lanes contain molecular size standard (100 bp ladder). |
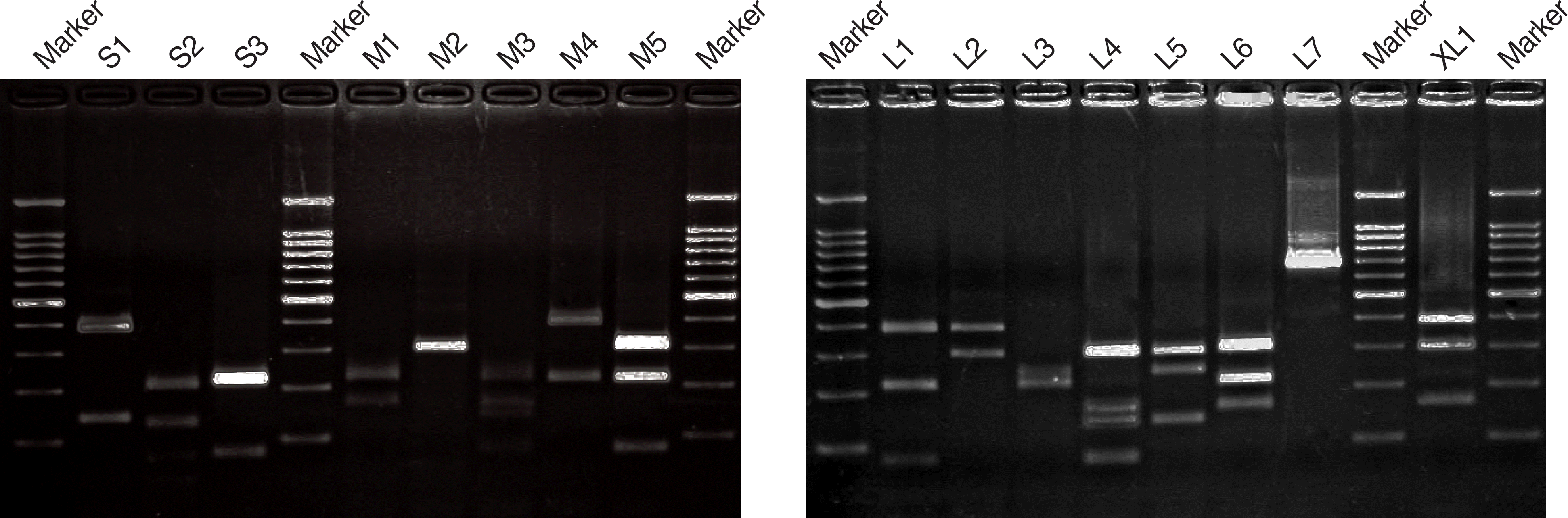 | Fig. 2.Agarose gel electrophoresis of PCR-amplified coa genes digested with the restriction endonuclease AluI from representative strains of S. aureus. Marker Lanes contain molecular size standard (100 bp ladder). |
Table 1.
A number of 120 S. aureus blood isolates according to RFLP patterns, staphylococcal toxic genes, mecA gene, and methicillin resistance
| PCR-amplified coa gene size∗ | RFLP pattern† | N | Toxin gene | mecA gene | MRSA (N=85) | MSSA (N=35) |
|---|---|---|---|---|---|---|
| S (N=6) | S1 | 1 | sec, tst | + | 1 | |
| S2 | 1 | - | + | 1 | ||
| tst | - | 1 | ||||
| S3 | 4 | sea, tst | - | 2 | ||
| sea, see, tst | - | 1 | ||||
| M (N=13) | M1 | 1 | - | - | 1 | |
| M2 | 1 | - | - | 1 | ||
| M3 | 1 | - | - | 1 | ||
| M4 | 2 | - | - | 2 | ||
| - | - | 1 | ||||
| M5 | 8 | sea | - | 3 | ||
| see | - | 1 | ||||
| sea, see | - | 3 | ||||
| L (N=97) | L1 | 37 | - | - | 1 | |
| - | + | 4 | ||||
| sea | + | 16 | ||||
| sea, see | + | 16 | ||||
| L2 | 3 | - | + | 2 | ||
| sec, tst | + | 1 | ||||
| L3 | 3 | sea | - | 1 | ||
| sea, see | - | 2 | ||||
| L4 | 1 | sec, tst | + | 1 | ||
| L5 | 46 | - | - | 1 | ||
| sea | - | 1 | ||||
| sea, see | - | 1 | ||||
| - | + | 2 | ||||
| tst | + | 6 | ||||
| sea, sec | + | 1 | ||||
| sec, tst | + | 32 | ||||
| sea, sec, tst | + | 2 | ||||
| L6 | 4 | - | - | 4 | ||
| L7 | 3 | - | - | 1 | ||
| sec | - | 2 | ||||
| XL (N=4) | XL1 | 4 | - | - | 4 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download