Abstract
Background
We intended to evaluate the diagnostic usefulness of a multiplex reverse transcriptase-PCR (mRT-PCR) assay kit under dual priming oligonucleotide system (DPO) for the childhood acute respiratory tract infections.
Methods
Two hundred nasopharyngeal aspirates were taken from children ≤5 yr old admitted due to acute respiratory infections in 2004. Direct fluorescent antibody (FA) assays were performed with fresh specimens; then, mRT-PCRs for the detection of 12 respiratory viruses (Seeplex RV detection kit, SeeGene, Seoul, Korea) were tested with frozen specimens.
Results
FA assays for five common respiratory viruses showed positive results in 66 patients (33.0%), while mRT-PCR detected causative viruses in 112 patients (56.0%), including 16 co-infected cases (8.0%). A total of 129 viruses were identified: respiratory syncytial virus A/B (38.0%/7.8%), influenza virus A/B (10.1%/5.4%), parainfluenza virus 1/2/3 (7.0%/3.1%/7.8%), coronavirus 229E or NL63 (6.2%), human metapneumovirus (4.7%), adenovirus (4.7%), rhinovirus (3.9%), and coronavirus OC43 (1.6%).
Go to : 
REFERENCES
1.Scheltinga SA., Templeton KE., Beersma MF., Claas EC. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J Clin Virol. 2005. 33:306–11.

2.Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A, et al. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol. 2006. 78:938–49.

3.van Woensel JB., van Aalderen WM., Kimpen JL. Viral lower respiratory tract infection in infants and young children. BMJ. 2003. 327:36–40.

4.Chun JY., Kim KJ., Hwang IT., Kim YJ., Lee DH., Lee IK, et al. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007. 35:e40.

5.Elnifro EM., Ashshi AM., Cooper RJ., Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000. 13:559–70.

6.Chung JY., Han TH., Kim BE., Kim CK., Kim SW., Hwang ES. Human metapneumovirus infection in hospitalized children with acute respiratory disease in Korea. J Korean Med Sci. 2006. 21:83842.

7.Choi EH., Lee HJ., Kim SJ., Eun BW., Kim NH., Lee JA, et al. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin Infect Dis. 2006. 43:585–92.

8.Weigl JA., Puppe W., Grondahl B., Schmitt HJ. Epidemiological investigation of nine respiratory pathogens in hospitalized children in Germany using multiplex reverse-transcriptase polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 2000. 19:336–43.

9.Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J, et al. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005. 126:53–63.

10.Kim YK., Lee HJ. Human metapneumovirus-associated lower respiratory tract infections in korean infants and young children. Pediatr Infect Dis J. 2005. 24:1111–2.

11.Kim SH., Huh JH., Bae SY., Kim JS., Yoon SY., Lim CS, et al. Epidemiology of respiratory viral infection in 2004-2006. Korean J Lab Med. 2006. 26:351–7. (김선형, 허지훈, 배숙영, 김장수, 윤수영, 임채승등. 2004-2006년의 호흡기 바이러스 감염의 역학. 대한진단검사의학회지 2006;26: 351-7.).

12.Han TH., Chung JY., Kim SW., Hwang ES. Human Coronavirus-NL63 infections in Korean children, 2004-2006. J Clin Virol. 2007. 38:27–31.

13.Sullender WM. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev. 2000. 13:1–15.

14.Wertz GW., Moudy RM. Antigenic and genetic variation in human respiratory syncytial virus. Pediatr Infect Dis J. 2004. 23(S):S19–24.

15.Morgan LA., Routledge EG., Willcocks MM., Samson AC., Scott R., Toms GL. Strain variation of respiratory syncytial virus. J Gen Virol. 1987. 68:2781–8.

Go to : 
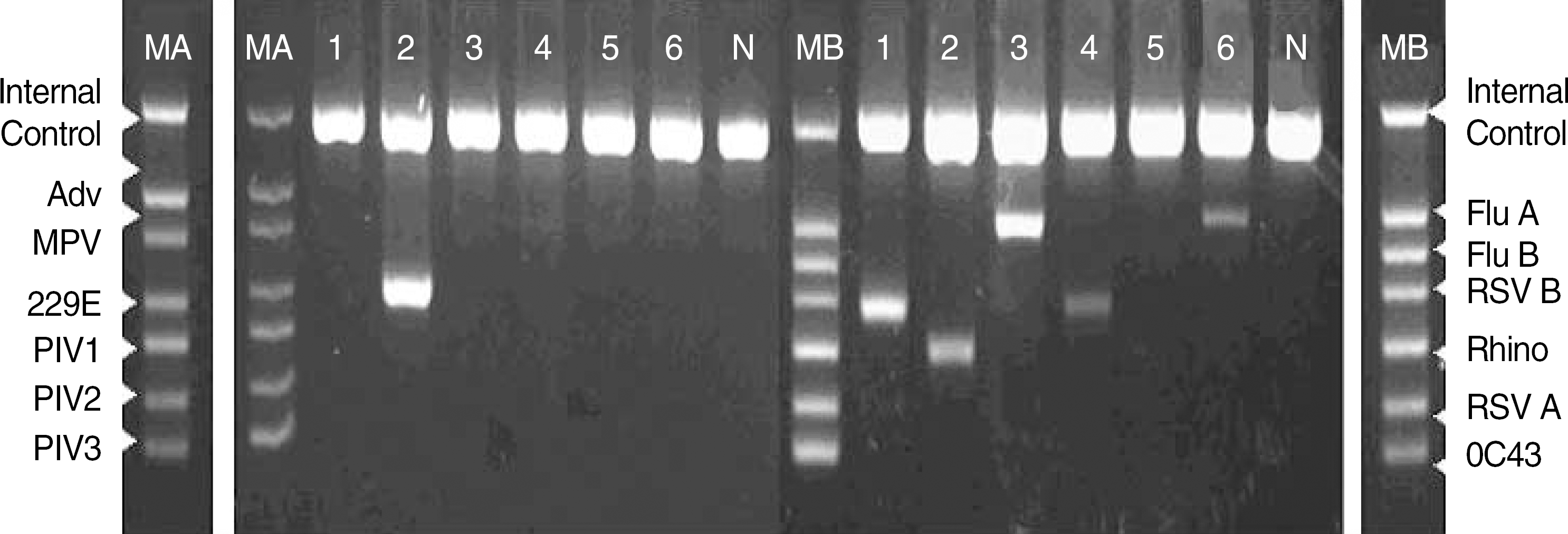 | Fig. 1.Two sets (A set in the left and B set in the right) of mRT-PCR using dual priming oligonucleotide systems in 6 clinical specimens. Lane MA, Size marker of A set; MB, Size marker of B; N, negative control; 1-6, Patients 1-6; Patient 1, Positive for RSV B; Patient 2, coronavirus 229E/NL63 and rhinovirus; Patient 3, influenza A; Patient 4, RSV B, Patient 5, negative; Patient 6, influenza A. Each lane presents internal control band (719 bp). Adv, Adenovirus (534 bp); MPV, Human metapneumovirus (469 bp); 229E, Corovirus 229E/NL63 (375 bp); PIV1, Parainfluenza virus 1 (324 bp); PIV2, Parainfluenza virus 2 (264 bp); PIV3, Parainfluenza virus 3 (219 bp); Flu A, Influenza A virus (516 bp); Flu B, Influenza B virus (455 bp); RSV B, Respiratory syncytial virus B (391 bp); Rhino, Human rhinovirus A (340 bp); RSV A, Respiratory syncytial virus A (273 bp); OC43, Coronavirus OC43 (231 bp). Abbreviation: mRT-PCR, multiple reverse transcriptase PCR. |
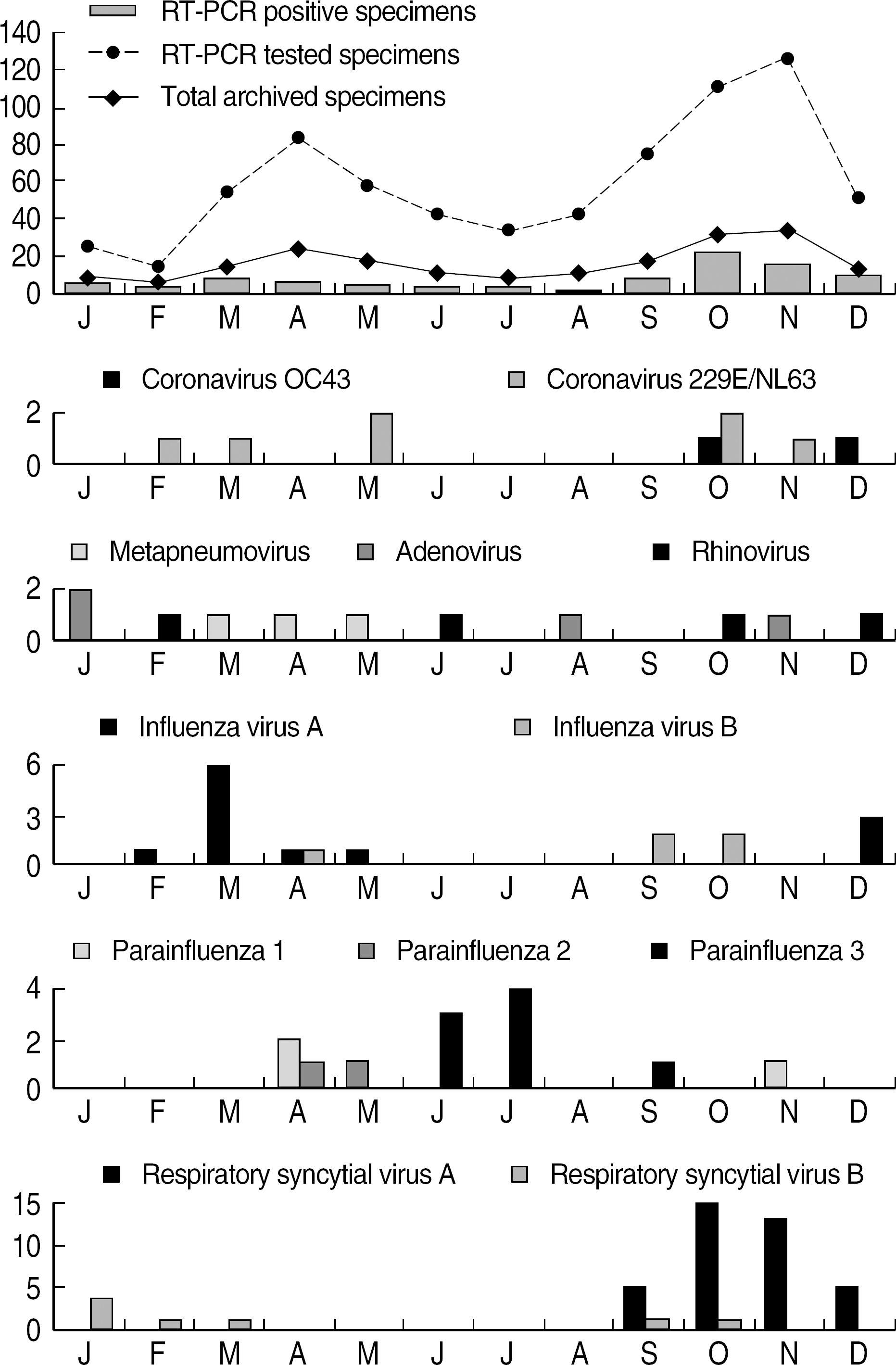 | Fig. 2.Monthly distribution of 12 respiratory viruses isolated from children with acute respiratory tract infections in 2004 (Co-infected cases were excluded). Abbreviation: mRT-PCR, multiple reverse transcriptase PCR. |
Table 1.
Viruses identified in 200 nasopharyngeal aspirates obtained from children with acute respiratory tract infections by mRT-PCR and fluorescent antibody assays
| Virus identified | N (%) of positive specimens, by the indicated method of detection (N=200) | Concordance of mRT-PCR and fluorescent Ab assay | ||||
|---|---|---|---|---|---|---|
| mRT-PCR | Fluorescent Ab assay | Both (+) | PCR (+) Ab (–) | PCR (–) Ab (+) | Concordance (%)∗ | |
| RSV A | 49 (24.5) | 45 (22.5)† | 43 | 6 | 2 | 84.3 |
| RSV B | 10 (5.0) | 2 (1.0)† | 2 | 8 | 0 | 20.0 |
| Influenza virus A | 13 (6.5) | 6 (3.0) | 5 | 8 | 1 | 35.7 |
| Influenza virus B | 7 (3.5) | 2 (1.0) | 0 | 7 | 2 | 0 |
| Parainfluenza virus 1 | 9 (4.5) | 2 (1.0)† | 2 | 7 | 0 | 22.2 |
| Parainfluenza virus 2 | 4 (2.0) | 0† | 0 | 4 | 0 | 0 |
| Parainfluenza virus 3 | 10 (5.0) | 7 (3.5)† | 7 | 3 | 0 | 70.0 |
| Adenovirus | 6 (3.0) | 2 (1.0) | 2 | 4 | 0 | 33.3 |
| Coronavirus 229E/NL63 | 8 (4.0) | Not tested | ||||
| Coronavirus OC43 | 2 (1.0) | Not tested | ||||
| Human metapneumovirus | 6 (3.0) | Not tested | ||||
| Rhinovirus | 5 (2.5) | Not tested | ||||
| Total | 129 (56.0)† | 66 (33.0) | 61 | 47 | 5 | 54.0 |
∗ Corcordance rate (%)=(N of positive results by both of the two methods∗100)/(N of positive results by both of the two methods + N of positive results in multiplex RT-PCR only+N of positive results in fluorescent Ab assay only);
Table 2.
Disease entities in 200 children with acute respiratory tract infections and their causative agents detected by mRT-PCR∗




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download