Abstract
Background
Many methicillin-resistant Staphylococcus aureus (MRSA) isolates in Korea possess a specific profile of staphylococcal enterotoxins in that the toxic shock syndrome toxin gene (tst) coexists with the staphylococcal enterotoxin C gene (sec). Because the analysis of staphylococcal cassette chromosome mec (SCCmec), a mobile genetic element mecA gene encoding methicillin resistance, showed that majority of these are SCCmec type II, these MRSA isolates with tst and sec may be genetically related with each other. This study was performed to investigate the genetic relatedness of tst-and sec-harboring MRSA strains isolated in Korea by using pulsed-field gel electrophoresis (PFGE).
Methods
A total of 59 strains of MRSA isolates of SCCmec type II possessing tst and sec were selected for PFGE and phylogenetic analyses. These isolates were collected from 13 health care facilities during nationwide surveillance of antimicrobial resistance in 2002.
Results
The 59 MRSA isolates were clustered into 11 PFGE types, including one major group of 26 strains (44.1%) isolated from 7 healthcare facilities. Seven PFGE types contained 2 or more isolates each, comprising 55 isolates in total.
Conclusions
Most of SCCmec type II MRSA isolates containing tst and sec showed closely related PFGE patterns. Moreover, MRSA isolates collected from different healthcare facilities showed identical PFGE patterns. These findings suggested a clonal spread of MRSA strains possessing tst and sec in Korean hospitals.
Go to : 
References
1. Chong Y, Lee K. Present situation of antimicrobial resistance in Korea. J Infect Chemother. 2000; 6:189–95.

2. Hong SG, Yong DE, Lee KW, Kim EC, Lee WK, Jeong SH, et al. Antimicrobial resistance of clinically important bacteria isolated from hospitals located in representative provinces of Korea. Korean J Clin Microbiol. 2003; 6:29–36.
3. Hong SG, Lee JW, Yong DE, Kim EC, Jeong SH, Park YJ, et al. Antimicrobial resistance of clinically important bacteria isolated from 12 hospitals in Korea. Korean J Clin Microbiol. 2004; 7:171–7.
4. Lee HM, Yong DE, Lee KW, Hong SG, Kim EC, Jeong SH, et al. Antimicrobial resistance of clinically important bacteria isolated from 12 hospitals in Korea in 2004. Korean J Clin Microbiol. 2005; 8:66–73.
5. Kim JS, Kim HS, Song WK, Cho HC, Lee KM, Kim EC. Antimicrobial resistance profiles of Staphylococcus aureus isolated in 13 Korean hospitals. Korean J Lab Med. 2004; 24:223–9.
6. Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997; 10:781–91.

7. Oliveira DC, Tomasz A, de Lencastre H. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002; 2:180–9.
8. Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006; 193:172–9.
9. Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, et al. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006; 50:1001–12.
10. McCormick JK, Yarwood JM, Schlievert PM. Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol. 2001; 55:77–104.

11. Chini V, Dimitracopoulos G, Spiliopoulou I. Occurrence of the ente-rotoxin gene cluster and the toxic shock syndrome toxin 1 gene among clinical isolates of methicillin-resistant Staphylococcus aureus is related to clonal type and agr group. J Clin Microbiol. 2006; 44:1881–3.
12. Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital-and community-associated strains of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2006; 193:1495–503.
13. Tsen HY, Yu GK, Hu HH. Comparison of type A enterotoxigenic Staphylococcus aureus strains isolated from geographically far distant locations by pulsed field gel electrophoresis. J Appl Microbiol. 1997; 82:485–93.
14. Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001; 3:585–94.

15. Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002; 70:4987–96.
16. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003; 41:1574–85.
17. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003; 41:5113–20.
18. Kim JS, Song W, Kim HS, Cho HC, Lee KM, Choi MS, et al. Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn Microbiol Infect Dis. 2006; 56:289–95.
19. Durand G, Bes M, Meugnier H, Enright MC, Forey F, Liassine N, et al. Detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in france. J Clin Microbiol. 2006; 44:847–53.
20. Nishi J, Yoshinaga M, Miyanohara H, Kawahara M, Kawabata M, Motoya T, et al. An epidemiologic survey of methicillin-resistant Staphylococcus aureus by combined use of mec-HVR genotyping and toxin genotyping in a university hospital in Japan. Infect Control Hosp Epidemiol. 2002; 23:506–10.
21. Soo Ko K, Kim YS, Song JH, Yeom JS, Lee H, Jung SI, et al. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob Agents Chemother. 2005; 49:3583–5.
22. Soo Ko K, Peck KR, Sup Oh W, Lee NY, Hiramatsu K, Song JH. Genetic differentiation of methicillin-resistant Staphylococcus aureus strains from Korea and Japan. Microb Drug Resist. 2005; 11:279–86.
23. Woo JH, Park KY, Chung DR, Kim EO, Kim YS, Ryu J. Molecular epidemiologic surveillance of TSST-1 strains among pathogenic Staphylococcus aureus. Korean J Infect Dis. 1997; 29:463–7.
24. Banks MC, Kamel NS, Zabriskie JB, Larone DH, Ursea D, Posnett DN. Staphylococcus aureus express unique superantigens depending on the tissue source. J Infect Dis. 2003; 187:77–86.
25. Fowler VG Jr, Justice A, Moore C, Benjamin DK Jr, Woods CW, Campbell S, et al. Risk factors for hematogenous complications of intravascular catheter-associated Staphylococcus aureus bacteremia. Clin Infect Dis. 2005; 40:695–703.
26. Ferry T, Thomas D, Genestier AL, Bes M, Lina G, Vandenesch F, et al. Comparative prevalence of superantigen genes in Staphylococcus aureus isolates causing sepsis with and without septic shock. Clin Infect Dis. 2005; 41:771–7.
27. Lehn N, Schaller E, Wagner H, Kronke M. Frequency of toxic shock syndrome toxin- and enterotoxin-producing clinical isolates of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1995; 14:43–6.
28. Schmitz FJ, MacKenzie CR, Geisel R, Wagner S, Idel H, Verhoef J, et al. Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin sensitive Staphylococcus aureus strains. Eur J Epidemiol. 1997; 13:699–708.
29. Shukla SK, Stemper ME, Ramaswamy SV, Conradt JM, Reich R, Graviss EA, et al. Molecular characteristics of nosocomial and native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J Clin Microbiol. 2004; 42:3752–7.
30. Wielders CL, Fluit AC, Brisse S, Verhoef J, Schmitz FJ. mecA gene is widely disseminated in Staphylococcus aureus population. J Clin Microbiol. 2002; 40:3970–5.
Go to : 
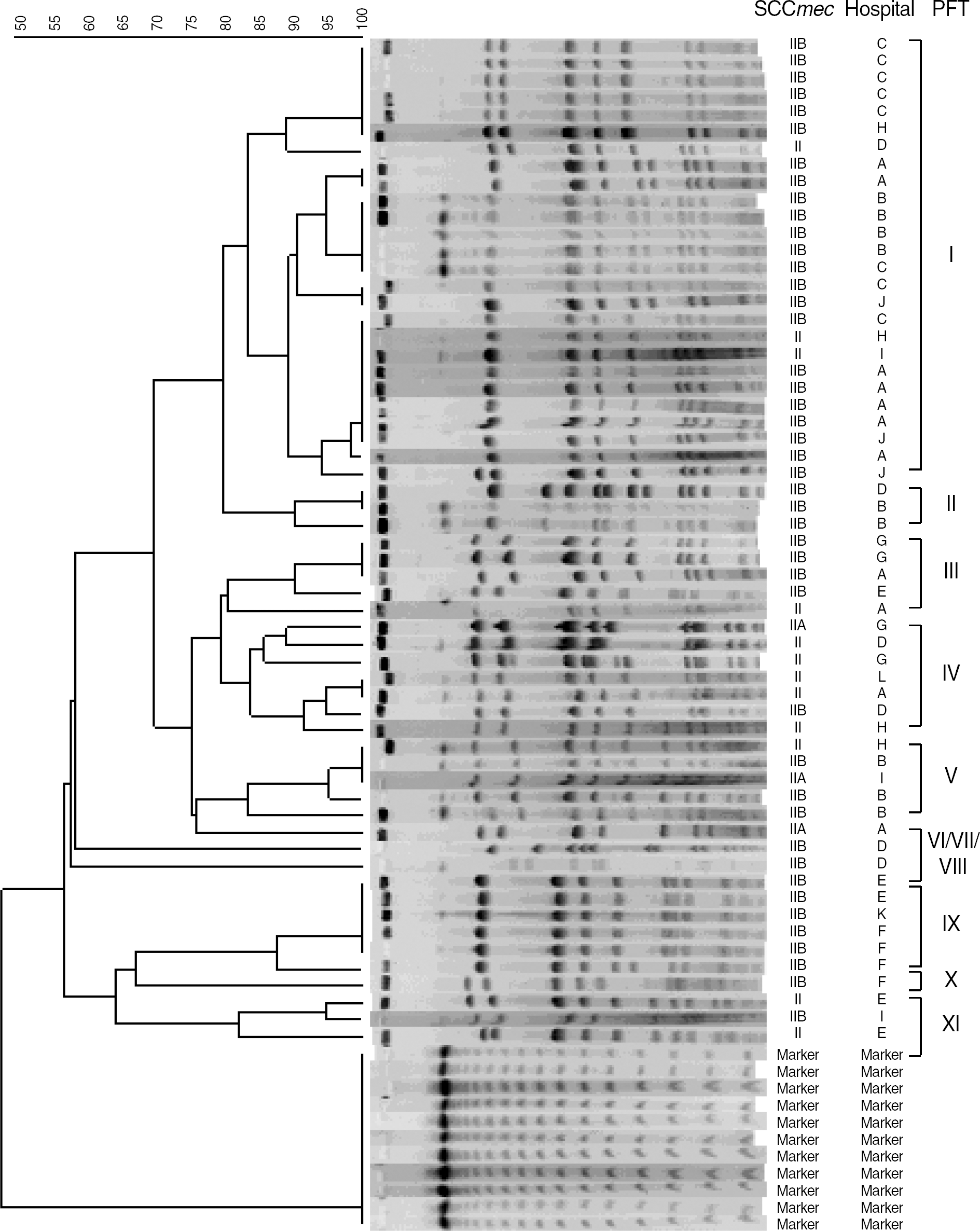 | Fig. 1.Dendrogram showing the PFGE patterns of SCCmec II isolates containing sec and tst. Also shown are the corresponding SCCmec types and 12 hospital origins.
Abbreviations: PFGE, pulsed-field gel electrophoresis; SCCmec, staphylococcal cassette chromosome mec; PFT, pulsed-field type.
|
Table 1.
Characteristics of PFGE types, SCCmec subtypes, and isolated hospitals of tst- and sec-positive methicillin-resistant Staphylococcus aureus isolates. PFGE types were classified by the similarity genetic coefficiency of 80%




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download