Abstract
Background
Enterovirus is a common cause of aseptic meningitis, respiratory disease and nonspecific febrile illness. The conventional methods for laboratory diagnosis of enterovirus infections have been virus culture and serotyping by an immunofluorecent test. We studied a new and more rapid approach for enterovirus detection in cerebrospinal fluid (CSF) by real-time nested PCR.
Methods
This study was performed on 50 CSF specimens from patients suspected of aseptic meningitis. Enterovirus was detected in CSF by PCRs for 3 different targets and real-time nested PCR. Enterovirus culture was also performed in 44 CSF specimens.
Go to : 
References
1. Rotbart HA, Sawyer MH, Fast S, Lewinski C, Murphy N, Keyser EF, et al. Diagnosis of enteroviral meningitis by using PCR with a colorimetric microwell detection assay. J Clin Microbiol. 1994; 32:2590–92.

2. Cherry JD, editor. Pediatric infectious disease. 3rd ed.Philadelphia: WB Saunders;1992. p. 1705–53.
3. White DO, Fenner FJ. Picoronaviridae. Fenner FJ, editor. Medical virology. 4th ed.San Diego: Academic Press;1994. p. 381–406.
4. Berlin LE, Rorabaugh ML, Heldrich F, Roberts K, Doran T, Modlin JF. Aseptic meningitis in infants <2 years of age: diagnosis and etiology. J Infect Dis. 1993; 168:888–92.
5. Modlin JF. Update on enterovirus infections in infants and children. Adv Pediatr Infect Dis. 1996; 12:155–80.
6. Hughes PJ, North C, Minor PD, Stanway G. The complete nucleotide sequence of coxsackievirus A21. J Gen Virol. 1989; 70:2943–52.

7. Hyypia T, Auvinen P, Maaronen M. Polymerase chain reaction for human picornaviruses. J Gen Virol. 1989; 70:3261–8.

8. Rotbart HA. Diagnosis of enteroviral meningitis with the polymerase chain reaction. J Pediatr. 1990; 117:85–9.

9. Schlesinger Y, Sawyer MH, Storch GA. Enteroviral meningitis in infancy: potential role for polymerase chain reaction in patient management. Pediatrics. 1994; 94:157–62.

10. Lee KM, Park SY, Kang HJ, Lee EH. Relation of sampling time to the detection of enteroviral RNA in cerebrospinal fluid from the patients with aseptic meningitis. Korean J Pediatr Infect Dis. 1996; 3:163–7.

11. Watkins-Riedel T, Woegerbauer M, Hollemann D, Hufnagl P. Rapid diagnosis of enterovirus infections by real-time PCR on the Light-Cycler using the TaqMan format. Diagn Microbiol Infect Dis. 2002; 42:99–105.

12. Oh SH, Lee MS, Kang JH, Kim CW, Park JY, Son YM, et al. Report of nationwide epidemiology of aseptic meningitis outbreak in 1993 in Korea. J Korean Pediatr Soc. 1996; 39:42–52.
13. Kim HK, Kang HJ, Lee KM. Echovirus type 9 infection in an epidemic of aseptic meningitis in 1993. Korean J Clin Path. 1994; 14:185–92.
14. Cho JY, Kim HJ, Jeong GY, Bang JK, Lee DP. Epidemic aseptic meningitis in 1993. J Korean Pediatr Soc. 1995; 38:901–6.
15. Kwon OS, Park SY, Lee KL, Kim WY, Jeong WJ, Ma SH. Epidemics of ascetic meningitis in kyoungsangnamdo from May to August, 1996. Korean J Pediatr Infect Dis. 1997; 4:97–105.

16. Park SY, Kwon OS, Kim WY, Jeong WJ, Ma SH, Lee KM. Epidemics of aseptic meningitis in kyoungsangnamdo from March to October, 1997. Korean J Pediatr Infect Dis. 1998; 5:104–14.

17. Peigue-Lafeuille H, Croquez N, Laurichesse H, Clavelou P, Aumaitre O, Schmidt J, et al. Enterovirus meningitis in adults in 1999–2000 and evaluation of clinical management. J Med Virol. 2002; 67:47–53.

18. Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997; 245:154–60.

19. Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997; 22:130–8.

20. Lee NY, Park HY, Lee YH, Seo JS. Clinical significance of HER2/neu gene quantification and MAGE assay in primary breast cancer. Korean J Lab Med. 2004; 24:432–8.
21. Seong MW, Kim JY, Ko HS, Hwang JM, Park SS. Mitochondrial DNA content and the MTND4 gene expression in leber's hereditary optic neuropathy. Korean J Lab Med. 2004; 24:439–45.
Go to : 
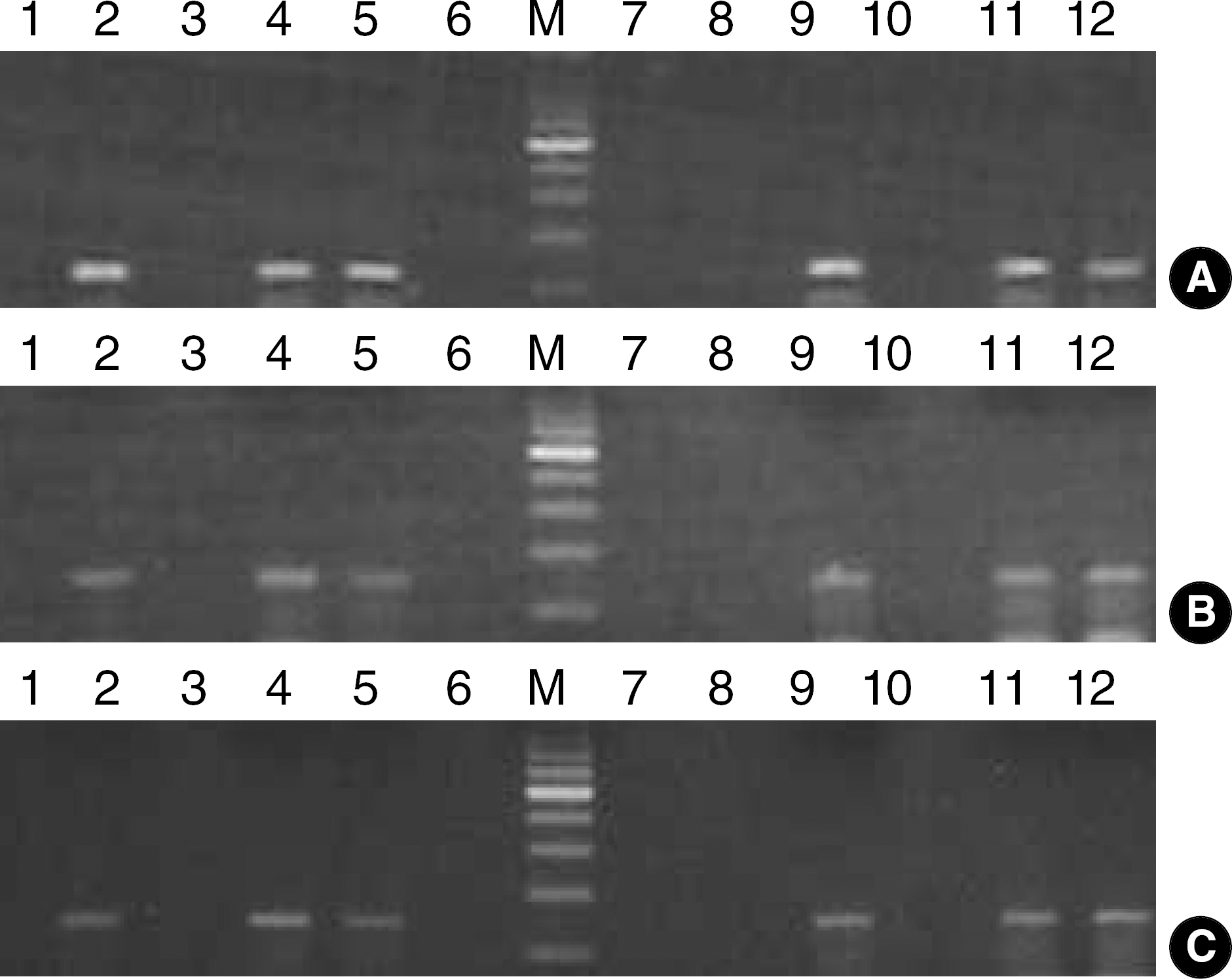 | Fig. 1.Detection of enterovirus by RT-PCR methods for 3 different targets using CSF specimens. (A) The amplified products of first method using EV1-F and EV1-R primers are 114 bp. (B) The amplified products of second method using EV2-F and EV2-R primers are 154 bp. (C) The amplified products of third method using EV3-F and EV3-R primers are 149 bp. Lane 1, negative control; lane 2, positive control; lane M, 100 bp DNA size marker; lane 3–12, specimens. |
Table 1.
Oligonucleotide primers used to detect enterovirus and reaction conditions
|
Methods |
PCR* | |
|---|---|---|
| 1 | Primers | EV1-F: 5′-ACA CGG ACA CCC AAA GTA GTC GGT TCC-3′ |
| EV1-R: 5′-TCC GGC CCC TGA ATG CGG CTA ATC C-3′ | ||
| Conditions Product size | 95°C 45 sec-58°C 45 sec-72°C 45 sec (35 cycles) 114 bp | |
| 2 | Primers | EV2-F: 5′-CCT CCG GCC CCT GAA TGC GGC TAA T-3′ |
| EV2-R: 5′-ATT GTC ACC ATA AGC AGC CA-3′ | ||
| Conditions Product size | 90°C 45 sec-46°C 45 sec-72°C 45 sec (37 cycles) 154 bp | |
| 3 | Primers | EV3-F: 5′-CCC CTG AAT GCG GCT AAT CC-3′ |
| EV3-R: 5′-CAA TTG TCA CCA TAA GCA GCC A-3′ | ||
| Conditions Product size | 95°C 45 sec-64.5°C 45 sec-72°C 45 sec (35 cycles) 149 bp | |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download