Abstract
Background
Recently, vancomycin-resistant enterococci (VRE) with the vanA genotype that are susceptible to teicoplanin have been described in Japan, Taiwan, and Korea. The investigators suggested three point mutations in the putative sensor domain of vanS or impairment of accessory proteins VanY and VanZ as an explanation for the VanB phenotype-vanA genotype VRE. In this study, we analyzed Tn1546-like elements to determine the molecular mechanisms responsible for the impaired glycopeptide resistance of clinical VRE isolates with VanB phenotype-vanA genotype from Korea.
Methods
From 2001 to 2004, 28 clinical isolates of Enterococcus faecium with VanB phenotype-vanA genotype were collected from 8 different university hospitals in diverse geographic areas in Korea. For structural analysis of Tn1546-like elements, PCR amplifications for internal regions of Tn1546 were performed. The purified PCR products were directly sequenced with an ABI Prism 3100 DNA sequencer.
Results
The sequence data of the vanS regulatory gene revealed that none of the isolates had any point mutations in this gene. All 28 isolates had a complete or incomplete deletion of vanY gene. Of these, 13 strains represented a complete deletion of vanZ, and 2 strains showed the deletion of nucleotides near the end point of vanX.
Go to : 
References
1. Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin & teicoplanin in Enterococcus faecium. N Engl J Med. 1988; 319:157–61.
2. Uttley AH, Collins CH, Naidoo J, George RC. Vancomycin-resistant enterococci. Lancet. 1988; 1:57–8.

3. Centers for Disease Control and Prevention. Nosocomial enterococci resistance to vancomycin-United States, 1989–1993. MMWR Morb Mortal Wkly Rep. 1993; 42:597–9.
4. National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004; 32:470–85.
5. Park JW, Kim YR, Shin WS, Kang MW, Han KJ, Shim SI. Susceptibility tests of vancomycin-resistant enterococci. Korean J Infect Dis. 1992; 24:133–8.
6. Shin JW, Yong D, Kim MS, Chang KH, Lee K, Kim JM, et al. Sudden increase of vancomycin-resistant enterococcal infections in a Korean tertiary care hospital: possible consequences of increased use of oral vancomycin. J Infect Chemother. 2003; 9:62–7.

7. Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000; 13:686–707.

8. Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993; 37:1563–71.

9. Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992; 36:2005–8.
10. Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997; 41:2016–8.
11. Fines M, Perichon B, Reynolds P, Sahm DF, Courvalin P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999; 43:2161–4.
12. McKessar SJ, Berry AM, Bell JM, Turnidge JD, Paton JC. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000; 44:3224–8.
13. Hashimoto Y, Tanimoto K, Ozawa Y, Murata T, Ike Y. Amino acid substitutions in the vanS sensor of the vanA-type vancomycin-resistant enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol Lett. 2000; 185:247–54.
14. Lauderdale TL, McDonald LC, Shiau YR, Chen PC, Wang HY, Lai JF, et al. Vancomycin-resistant enterococci from humans and retail chickens in Taiwan with unique VanB phenotype-vanA genotype incongruence. Antimicrob Agents Chemother. 2002; 46:525–7.
15. Eom JS, Hwang IS, Hwang BY, Lee JG, Lee YJ, Cheong HJ, et al. Emergence of vanA genotype vancomycin-resistant enterococci with low or moderate levels of teicoplanin resistance in Korea. J Clin Microbiol. 2004; 42:1785–6.
16. Lee WG, Huh JY, Cho SR, Lim YA. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob Agents Chemother. 2004; 48:1379–81.
17. Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 7th ed.Approved standard M7-A7. Wayne, Pa: Clinical and Laboratory Standards Institute;2006.
18. Murray BE, Singh KV, Heath JD, Sharma BR, Weinstock GM. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990; 28:2059–63.

19. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Sixteenth informational supplement, M2-A9 and M7-A7. Wayne, PA: Clinical and Laboratory Standards Institute;2006.
20. Arthur M, Molinas C, Depardieu F, Courvallin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993; 175:117–27.
21. Arthur M, Molinas C, Courvalin P. Sequence of the vanY gene required for production of a vancomycin-inducible D,D-carboxypeptidase in Enterococcus faecium BM4147. Gene. 1992; 120:111–4.
22. Arthur M, Molinas C, Courvallin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992; 174:2582–91.
Go to : 
 | Fig. 1.Dendrogram of E. faecium isolates with VanB phenotype-vanA. To produce the dendrogram, the banding patterns of E. faecium isolates were interpreted by Dice analysis and analysis by the unweighted pair group method with arithmetic averages. |
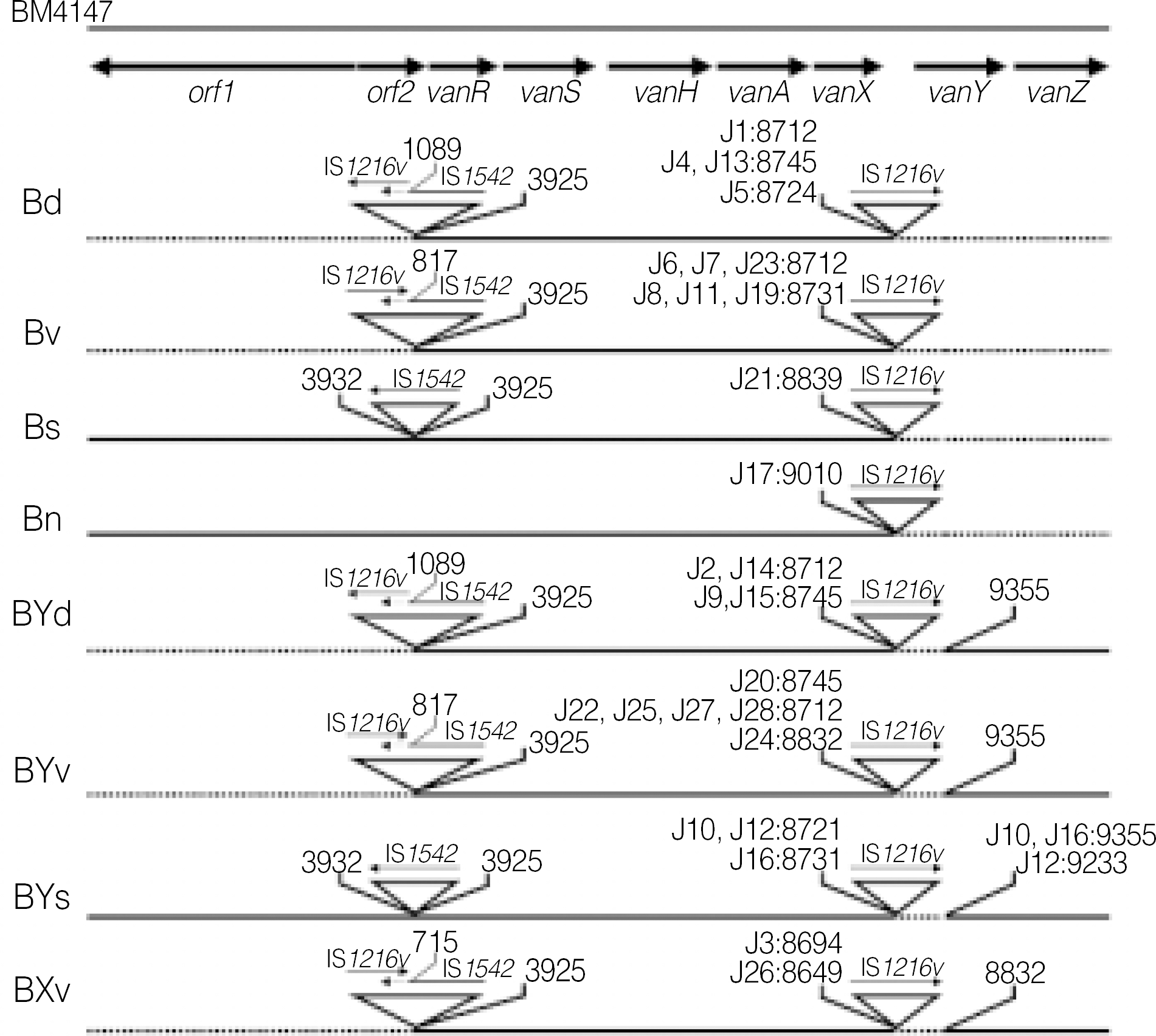 | Fig. 2.The genetic maps of Tn1546 types of E. faecium isolates with VanB phenotype-vanA genotype from Korean hospitals. The positions of genes and open reading frames (orf1 and orf2) and the direction of transcription are marked by dark arrows at the top. The inverted triangles represent IS elements. The positions of the first nucleotide upstream and the first nucleotide downstream from the IS insertion sites are depicted. Solid arrows indicate the transcriptional orientation of the inserted IS elements. Deletions are indicated by dotted lines. |
Table 1.
MICs of vancomycin and teicoplanin for clinical VRE isolates with VanB phenotype-vanA




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download