Abstract
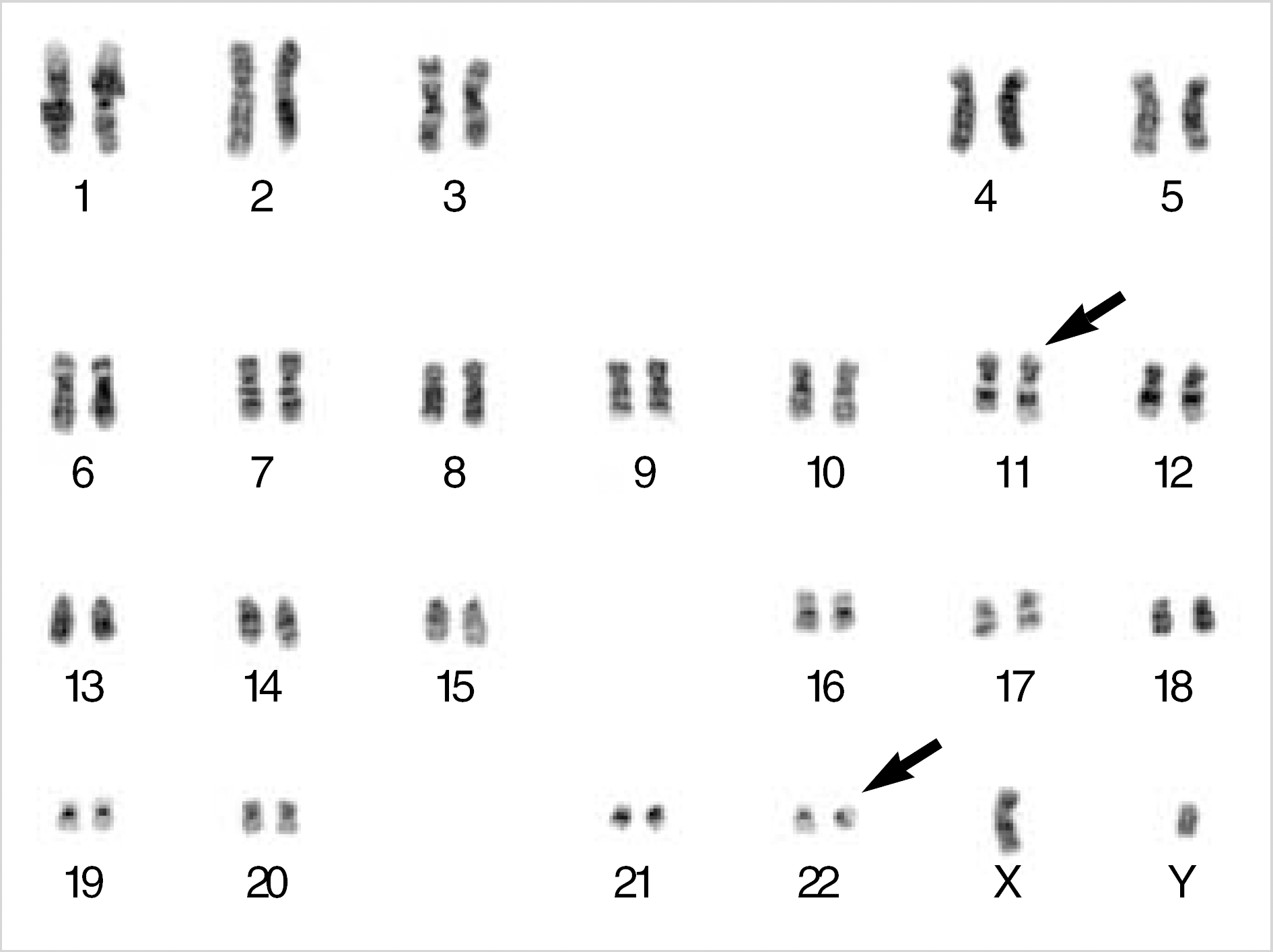
We report a case of chronic myelogenous leukemia displaying a variant Philadelphia translocation t(11;22)(q25;q11.2). Breakpoint 11q25 has not previously been reported. Reverse transcriptase polymerase chain reaction and fluorescence in-situ hybridization demonstrated the BCR/ABL rearrangement.
References
1. Heim S, Mitelman F, editors. Cancer cytogenetics. 2nd ed.New York: Wiley-Liss;1995. p. 40–3.
2. Anastasi J, Vardiman JW. Chronic myelogenous leukemia and the chronic myeloproliferative diseases. Knowles DM, editor. Neoplastic hematopathology. 2nd ed.Philadelphia: Lippincott Williams & Wilkins;2001. p. 1745–90.
3. Acar H, Stewart J, Boyd E, Connor MJ. Identification of variant translocations in chronic myeloid leukemia by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1997; 93:115–8.

4. Giere I, Migliorini AM, Bengio R, Arias D, Slavutsky I, Larripa I. Cytogenetic and molecular studies of variant Ph' translocations. Haematologica. 2000; 85:435–7.
5. Mitelman F, Johansson B, Mertens F. (Eds.),. Mitelman database of chromosome aberrations in cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman. (updated on May 22,. 2006.
6. Guillaume B, Ameye G, Libouton JM, Dierlamm J, Vaerman JL, Straetmans N, et al. Chronic myeloid leukemia with a rare variant Philadelphia translocation: t(9;22;21)(q34;q11;q22). Cancer Genet Cytogenet. 2000; 116:166–9.
7. Huntly BJ, Reid AG, Bench AJ, Campbell LJ, Telford N, Shepherd P, et al. Deletions of the derivative chromosome 9 occur at the time of the Philadelphia translocation and provide a powerful and independent prognostic indicator in chronic myeloid leukemia. Blood. 2001; 98:1732–8.

8. Cohen N, Amariglio N, Rechavi G, Trakhtenbrot L, Hardan I. Simultaneous detection of deletions of 9q and 22q in a subgroup of chronic myelocytic leukemia Philadelphia-positive patients by a novel probe. Cancer Genet Cytogenet. 2003; 141:89–90.

9. Lee YK, Kim YR, Lee DS, She CJ, Yoon SS, Park SY, et al. Clinical implication of the deletion status of ABL-BCR on derivative chromosome 9 in chronic myelogenous leukemia. Korean J Lab Med. 2002; 22:373–81.
10. Lee DS, Lee YS, Yun YS, Kim YR, Jeong SS, Lee YK, et al. A study on the incidence of ABL gene deletion on derivative chromosome 9 in chronic myelogenous leukemia by interphase fluorescence in situ hybridization and its association with disease progression. Genes Chromosomes Cancer. 2003; 37:291–9.
11. Yoong Y, VanDeWalker TJ, Carlson RO, Dewald GW, Tefferi A. Clinical correlates of submicroscopic deletions involving the ABL-BCR translocation region in chronic myeloid leukemia. Eur J Haematol. 2005; 74:124–7.
Fig. 2.
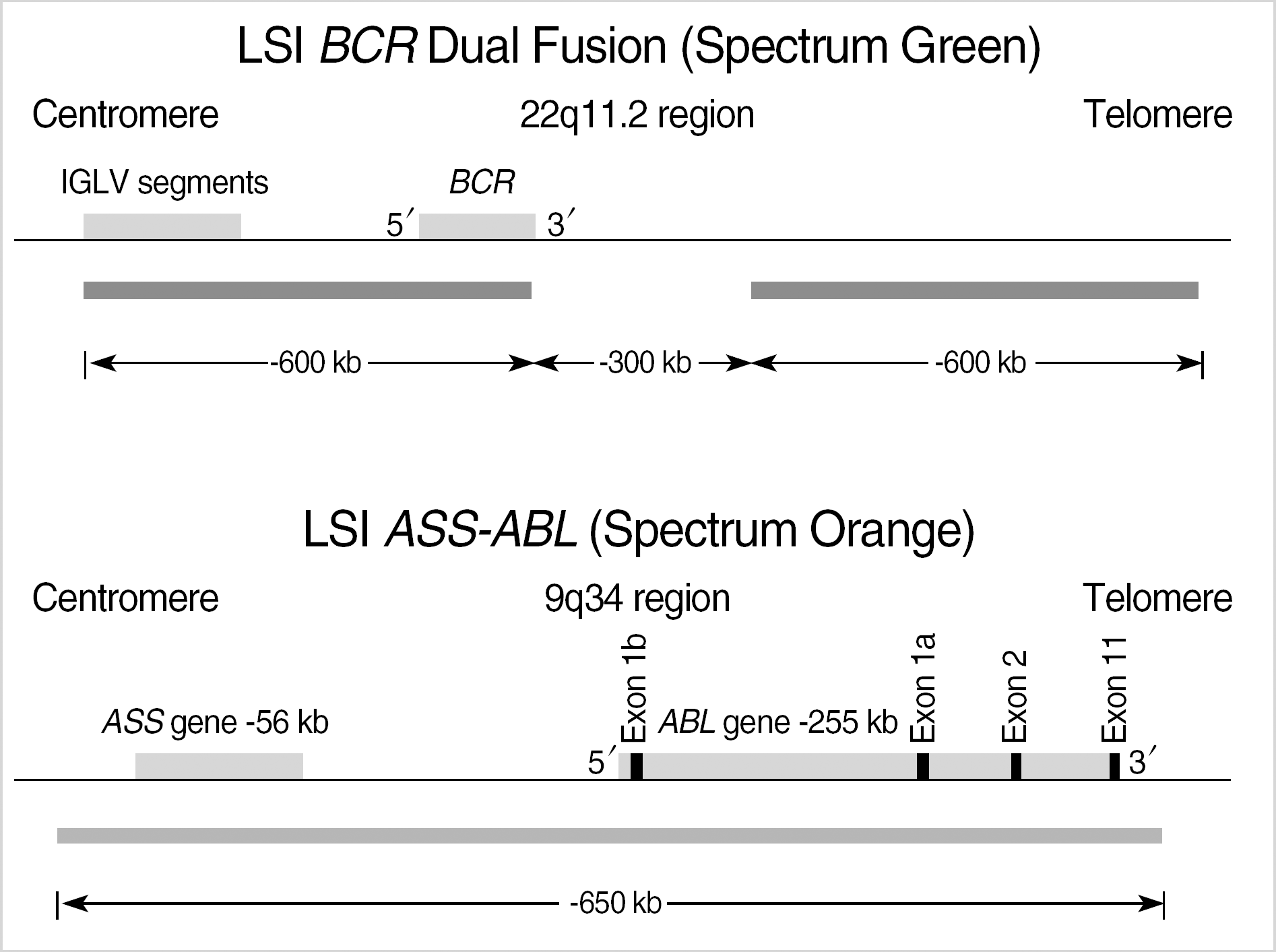
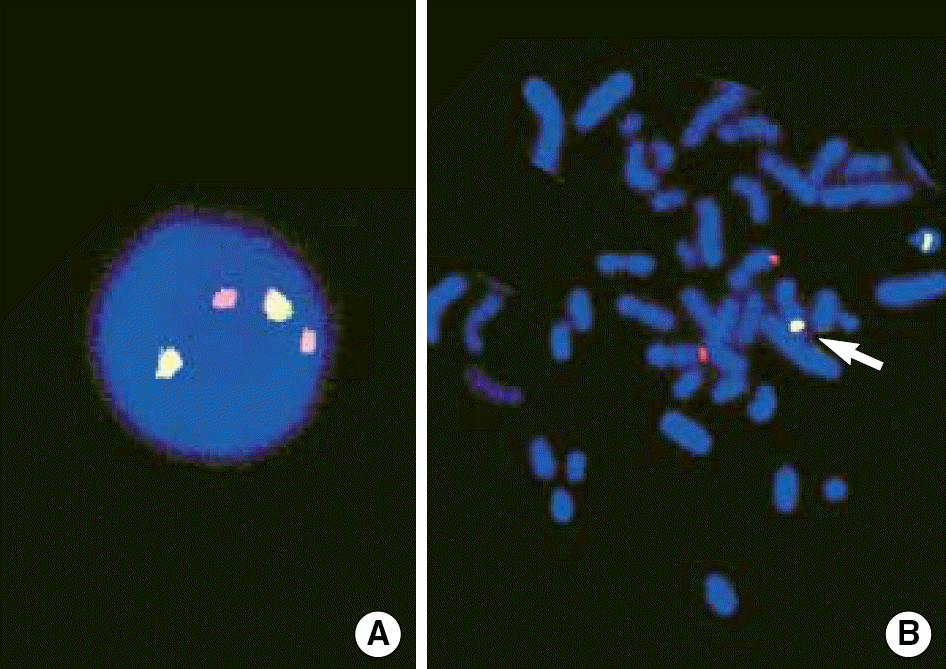
LSI BCR/ABL dual color, dual fusion probe (cited by http://www.vysis.com/AnalyticSpecificReagents(ASR)_59424.asp?PrintPage=true&ProdID=35–191032, Vysis, Downers Grove, IL, USA.). The BCR probe was labeled with a green fluorochrome (G) and the ASS-ABL probe was labeled with a orange fluorochrome (O). BCR/ABL rearrangements were detected as yellow fusion signals (F). The 2O2G was normal signal pattern. The 1O1G2F was typical pattern for BCR/ABL fusion. Atypical patterns of 1O1G1F, 1O2G1F and 2O1G1F are indicative of genomic deletions[9, 11].




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download