Abstract
Background
Two distinct types of fms-like tyrosine kinase 3 (FLT3) gene mutations have been identified in acute myeloid leukemia (AML): D835 and internal tandem duplication (ITD) mutations. These mutations are known to cause the proliferation of leukemic cells and inhibit the apoptosis of leukemic cells due to ligand-independent activation of their receptors. Therefore, the current study attempted to investigate the frequency of FLT3 gene mutations and their prognostic implications for AML in terms of treatment response, survival, and relapse.
Methods
Polymerase chain reaction (PCR) was performed to detect D835 and ITD mutations in 84 newly diagnosed AML patients from February 2001 to October 2004. Restriction fragment length polymorphism (RFLP) and direct sequencing were performed to analyze the D835 mutations. The results were examined based on a comparison with previously known prognostic factors, and the treatment outcomes analyzed according to the existence of the mutations in relation to the event free survival (EFS), overall survival (OS), and complete remission (CR) rates.
Results
D835 and IDT mutations were detected in 4.7% (4/84) and 19.0% (16/84), respectively, of the AML patients. The FLT3 gene mutations were not found to be associated with previously known prognostic factors, such as the WBC count, age, and cytogenetic risk group, but were associated with the lactate dehydrogenase levels. The EFS and OS rates were also significantly lower in the FLT3 gene mutation group, especially in AML with normal karyotypes.
Conclusions
FLT3 gene mutations were observed in 23.8% of AML patients and appeared to have a prognostic implication on patient survival. Accordingly, the presence of FLT3 gene mutations, which could be tested easily by using PCR/RFLP methods, should be investigated routinely at the time of diagnosis.
Go to : 
References
1. Hutchison RE, Davey FR. Leukocytic disorders. Henry JB, editor. Clinical diagnosis and management by laboratory methods. 20th ed.Philadelphia: WB Saunders;2001. p. 586–622.

2. Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999; 93:3074–80.
3. Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994; 84:3148–57.
4. Bergmann L, Miething C, Maurer U, Brieger J, Karakas T, Weidmann E, et al. High levels of Wilms' tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997; 90:1217–25.
5. Matthews W, Jordan CT, Wiegand GW, Pardoll D, Lemischka IR. A receptor tyrosine kinase specific to hematopoietic stem and progenitor cell-enriched populations. Cell. 1991; 65:1143–52.

6. Kottaridis PD, Gale RE, Linch DC. Flt3 mutations and leukaemia. Br J Haematol. 2003; 122:523–38.
7. Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003; 3:650–65.
8. Lisovsky M, Estrov Z, Zhang X, Consoli U, Sanchez-Williams G, Snell V, et al. Flt3 ligand stimulates proliferation and inhibits apoptosis of acute myeloid leukemia cells: regulation of Bcl-2 and Bax. Blood. 1996; 88:3987–97.
9. Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Genomic structure of human FLT3: implications for mutational analysis. Br J Haematol. 2001; 113:1076–7.
10. Shih LY, Huang CF, Wu JH, Wang PN, Lin TL, Dunn P, et al. Heterogeneous patterns of FLT3 Asp (835) mutations in relapsed de novo acute myeloid leukemia: a comparative analysis of 120 paired diagnostic and relapse bone marrow samples. Clin Cancer Res. 2004; 10:1326–32.
11. Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. Br J Haematol. 2001; 113:983–8.
12. Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood. 2001; 97:2434–9.
13. Thiede C, Steudel C, Mohr B, Schaich M, Schakel U, Platzbecker U, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002; 99:4326–35.
14. Murphy KM, Levis M, Hafez MJ, Geiger T, Cooper LC, Smith BD, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003; 5:96–102.
15. Shih LY, Kuo MC, Liang DC, Huang CF, Lin TL, Wu JH, et al. Internal tandem duplication and Asp835 mutations of the FMS-like tyrosine kinase 3 (FLT3) gene in acute promyelocytic leukemia. Cancer. 2003; 98:1206–16.
16. Tiesmeier J, Muller-Tidow C, Westermann A, Czwalinna A, Hoffmann M, Krauter J, et al. Evolution of FLT3-ITD and D835 activating point mutations in relapsing acute myeloid leukemia and response to salvage therapy. Leuk Res. 2004; 28:1069–74.
17. Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996; 10:1911–8.
18. Kiyoi H, Naoe T, Yokota S, Nakao M, Minami S, Kuriyama K, et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho). Leukemia. 1997; 11:1447–52.
19. Shih LY, Huang CF, Wu JH, Lin TL, Dunn P, Wang PN, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: a comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2002; 100:2387–92.
20. Abu-Duhier FM, Goodeve AC, Wilson GA, Gari MA, Peake IR, Rees DC, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol. 2000; 111:190–5.
21. Sohn YH, Chi HS, Joo MY, Huh HJ, Jang S, Park CJ. Investigation of Correlation of the recurrence of acute myolgenous leukemia and FLT3 mutation. Korean J Lab Med. 2004; 24:S402.
22. Yoo S, Shim EH, Park CJ, Chi HS. The frequency and clinical significance of FLT3 gene mutation in acute promyelocytic leukemia. Korean J Lab Med. 2003; 23:S174.
Go to : 
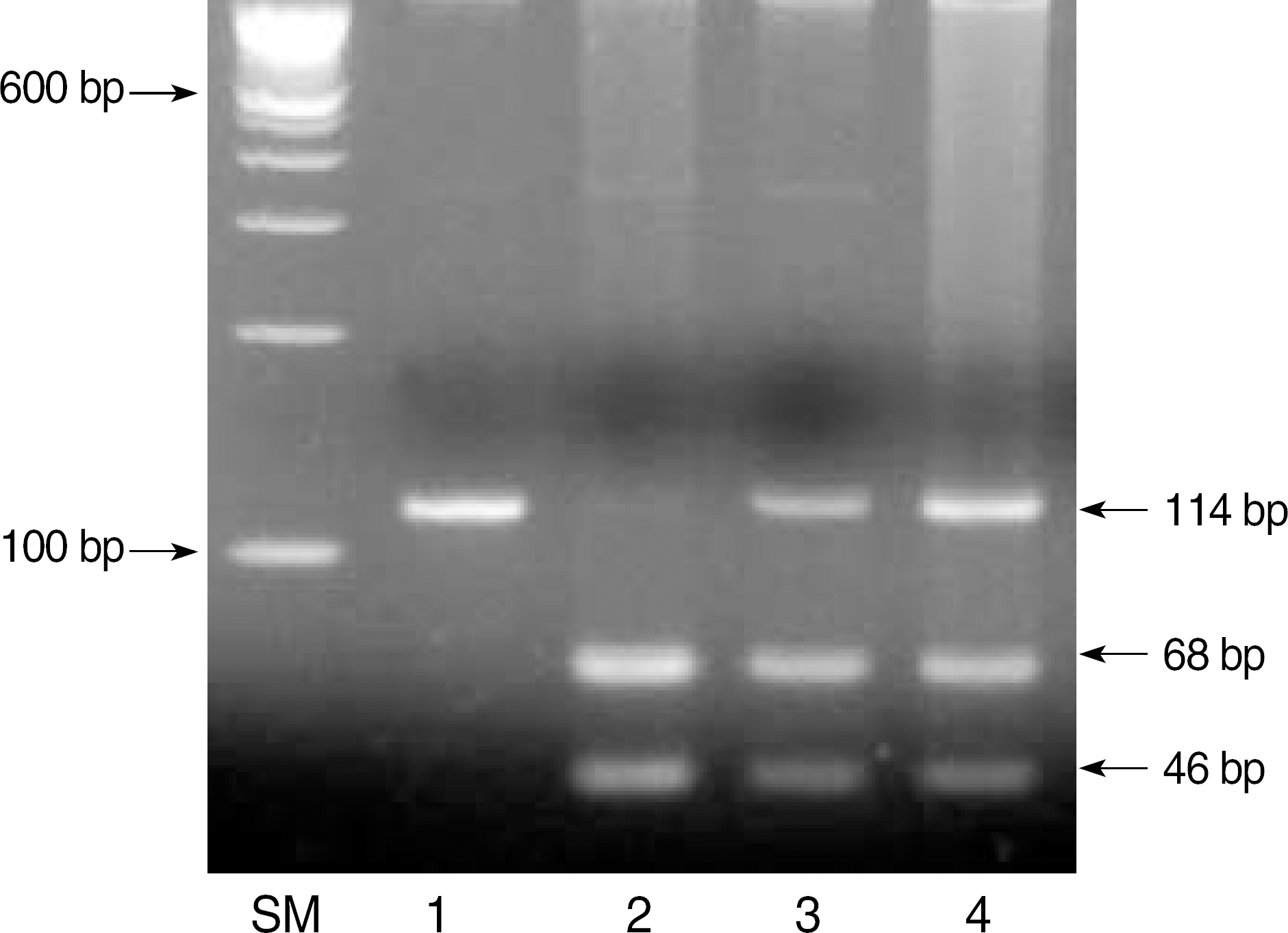 | Fig. 1.Results of D835 mutations in the FLT3 gene. The amplified product (lane 1) of wild type was digested to two bands (68 bp and 46 bp) by EcoRV in lane 2. When amplified products contained D835 mutations, undigested bands (114 bp) were visualized on low melting point agarose gel electrophoresis (lane 3 and 4). Lane SM, molecular size markers (100 bp DNA ladder). |
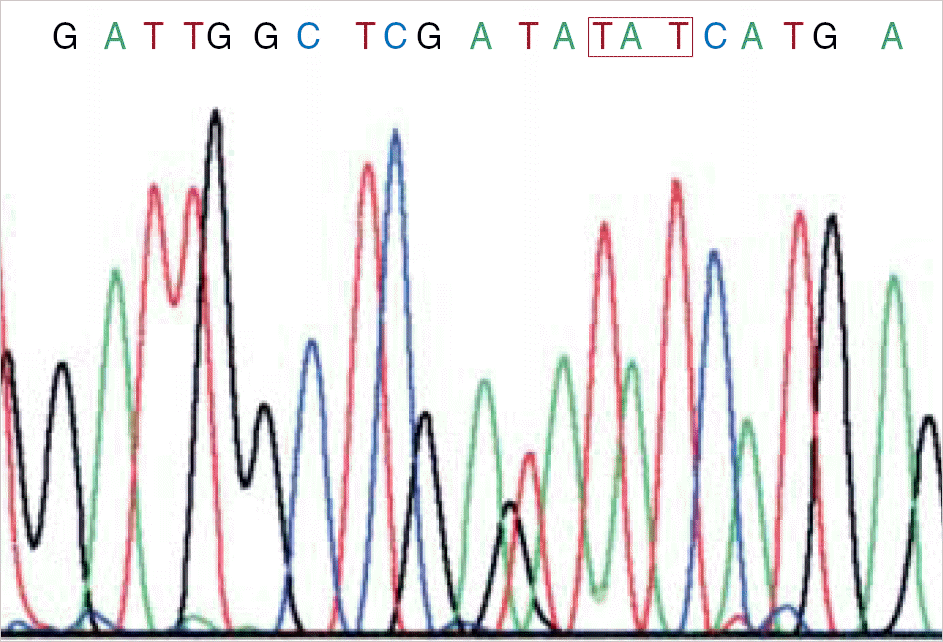 | Fig. 2.Sequence analysis in 3 patients with D835 mutations in the FLT3 gene. The results revealed that the first nucleotide G of codon 835 was substituted with T.
Abbreviation: FLT3, fms-like tyrosine kinase 3.
|
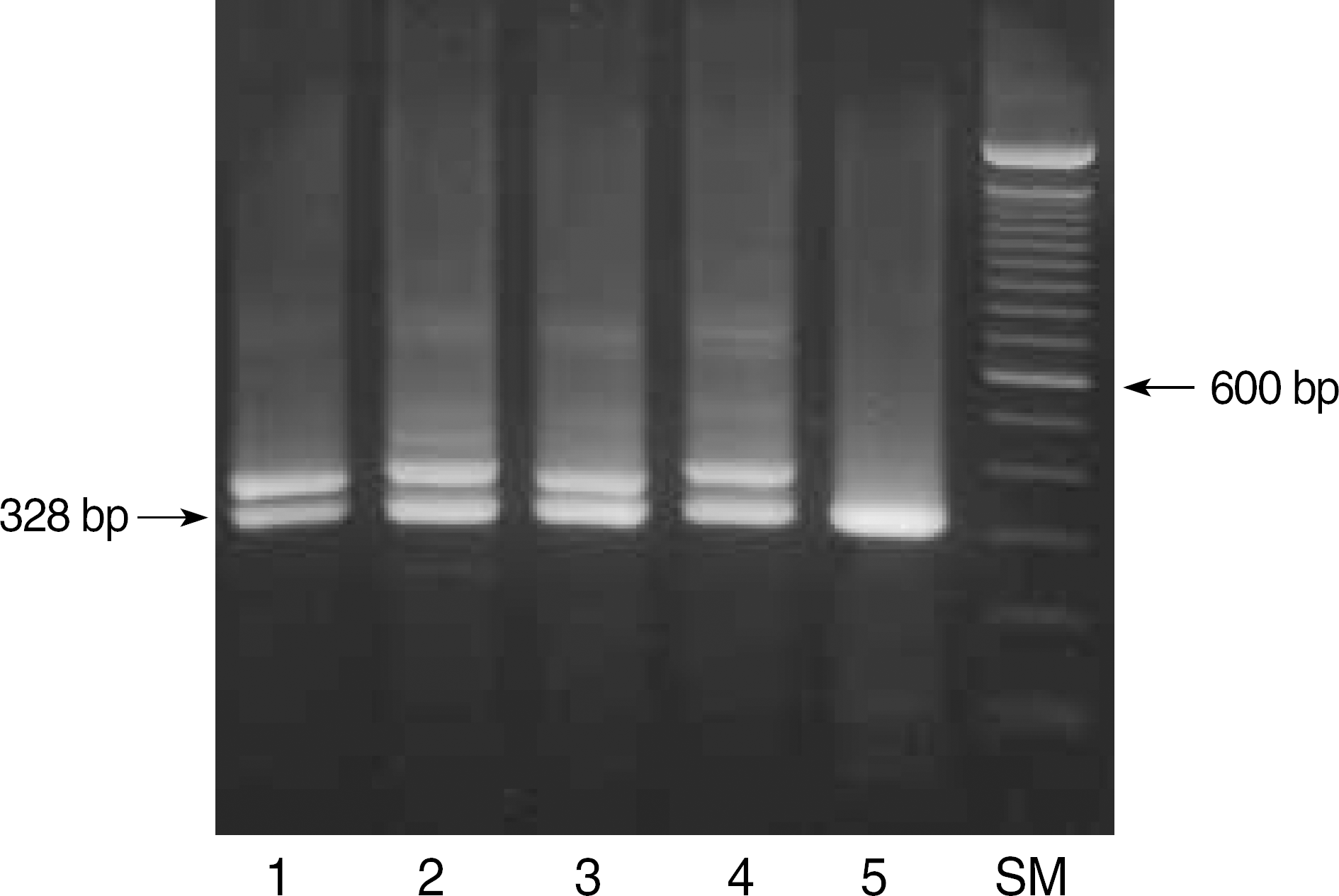 | Fig. 3.Results of ITD mutations in the FLT3 gene. The amplified product of wild type was shown in lane 5. When amplified products contained ITD mutations, another bands were visualized on low melting point agarose gel electrophoresis (lane 1–4). Lane SM, molecular size marker (100 bp DNA ladder). |
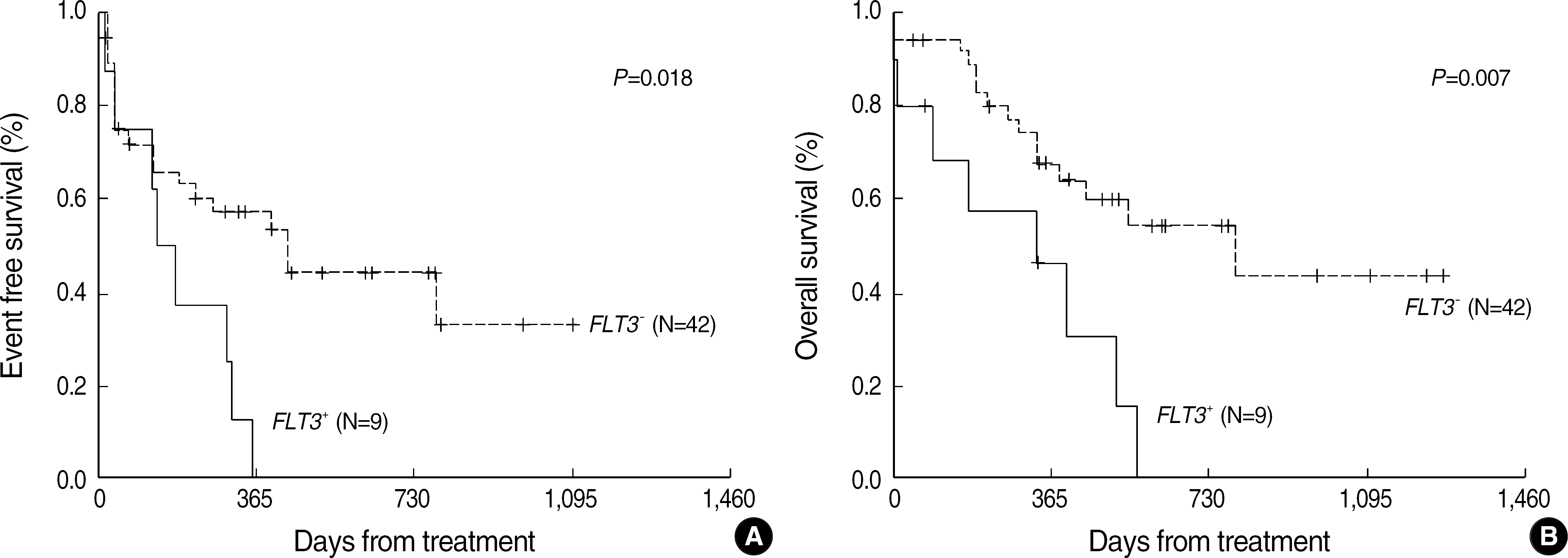 | Fig. 4.Event free (A) and overall (B) survival in patients with AML according to the FLT3 mutation status. |
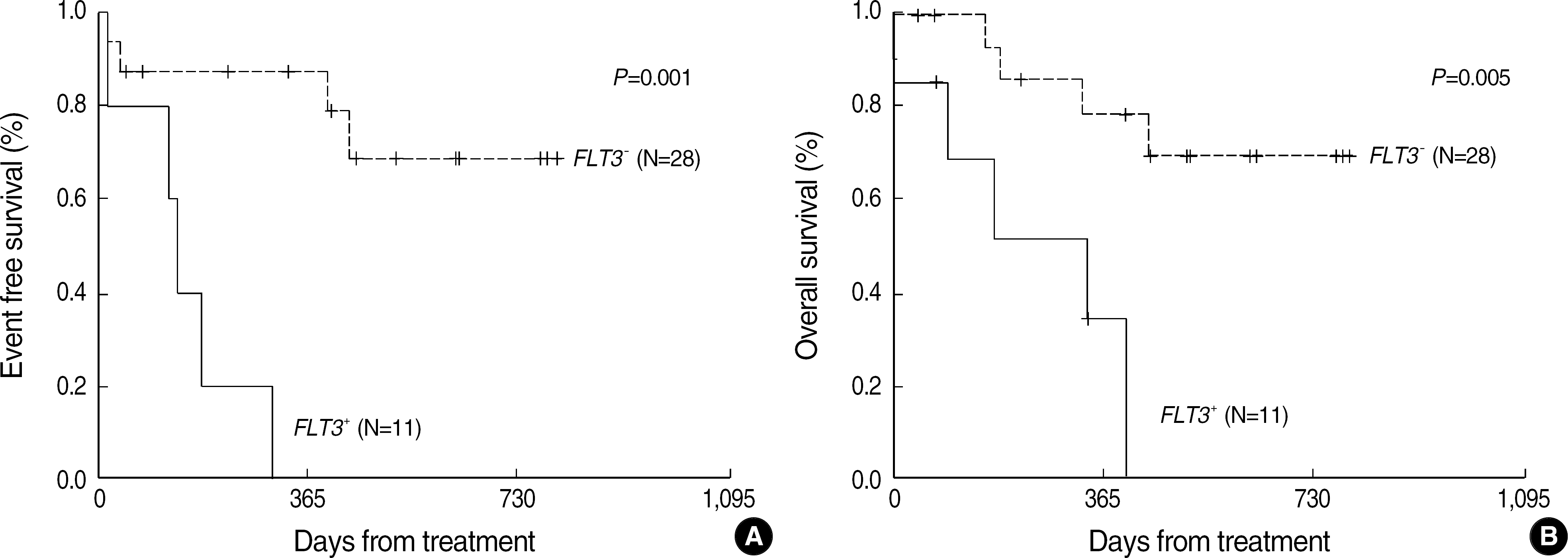 | Fig. 5.Event free (A) and overall (B) survival rate in patients with normal karyotype in AML according to the FLT3 mutation status. Abbreviations: AML, acute myeloid leukemia; FLT3, fms-like tyrosine kinase 3; ITD, internal tandem duplication; FLT3+, FLT3 mutation; FLT3-, no FLT3 mutation. |
Table 1.
Results of analysis for D835 and ITD mutations in the FLT3 gene
Table 2.
Relationship between known prognostic factors and the FLT3 mutations
| Prognostic factors | N of patient |
FLT3 mutations |
P value | |
|---|---|---|---|---|
| Positive | Negative | |||
| WBC (/μL) | ||||
| ≥50,000 | 22 | 5 | 17 | NS |
| <50,000 | 62 | 11 | 51 | |
| LD (U/L) | ||||
| ≥1,000 | 33 | 12 | 21 | 0.013 |
| <1,000 | 43 | 5 | 38 | |
| Age (years) | ||||
| ≥50 | 35 | 6 | 29 | NS |
| <50 | 49 | 14 | 35 | |
| Cytogenetic findings* | ||||
| Favorable group | 17 | 6 | 11 | NS |
| Intermediate group | 45 | 11 | 34 | |
| Unfavorable group | 15 | 3 | 12 | |
Table 3.
Comparison of survivals and complete remission rates excluding M3 according to the FLT3 mutation status
|
FLT3 mutations |
P value | ||
|---|---|---|---|
| Positive (N=9) | Negative (N=42) | ||
| EFS | |||
| Median (days) | 135 | 429 | 0.018 |
| 1 YEFS rate (%) | 0 | 57.0±8.5* | |
| OS | |||
| Median (days) | 336 | 793 | 0.007 |
| 1 YOS rate (%) | 45.7±16.6 | 67.8±8.0 | |
| CR | |||
| Rate (%) | 77.8 | 71.4 | NS |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download