Abstract
Background
Real-time polymerase chain reaction (PCR) is generally regarded as a very accurate and time-saving method, but it is expensive to run. We evaluated the reliability of an inexpensive and a researcher-friendly gel electrophoresis-based PCR method for the quantification of mRNA, and the results were compared with those obtained by real-time PCR.
Methods
We compared the results of relative quantification for MMP-1 measured by real-time PCR and by ethidium bromide stained-agarose gel electrophoresis after end-point PCR.
Results
There was significant but very weak correlation between real-time PCR and end-point PCR for relative quantification of MMP-1 (r=0.16, P<0.01).
Conclusions
Our results suggest that the use of the gel electrophoresis-based end-point PCR is inappropriate for quantifying mRNA. Therefore, in order to confirm the result of relative quantification by end-point PCR, the newly established real-time PCR method or northern hybridization should be applied.
References
1. Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987; 155:335–50.

2. Parker RM, Barnes NM. mRNA: detection by in situ and northern hybridization. Methods Mol Biol. 1999; 106:247–83.

3. Hod Y. A simplified ribonuclease protection assay. Biotechniques. 1992; 13:852–4.
4. Clarke PA, te Poele R, Wooster R, Workman P. Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential. Biochem Pharmacol. 2001; 62:1311–36.
5. Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000; 25:169–93.

6. Wang T, Brown MJ. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal Biochem. 1999; 269:198–201.

7. Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′→ 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991; 88:7276–80.
8. Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology. 1993; 11:1026–30.

9. Ovstebo R, Haug KB, Lande K, Kierulf P. PCR-based calibration curves for studies of quantitative gene expression in human monocytes: development and evaluation. Clin Chem. 2003; 49:425–32.
10. Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996; 6:986–94.

11. Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997; 22(130–1):134–8.

12. Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000; 285:194–204.

13. Heath PJ, Clendenning JB, Fujimoto BS, Schurr JM. Effect of bending strain on the torsion elastic constant of DNA. J Mol Biol. 1996; 260:718–30.

14. Schneeberger C, Speiser P, Kury F, Zeillinger R. Quantitative detection of reverse transcriptase-PCR products by means of a novel and sensitive DNA stain. PCR Method Appl. 1995; 4:234–8.

15. Hall LL, Bicknell GR, Primrose L, Pringle JH, Shaw JA, Furness PN. Reproducibility in the quantification of mRNA levels by RT-PCR-ELISA and RT competitive-PCR-ELISA. BioTechniques. 1998; 24:652–8.

16. Siddiqi AM, Jennings VM, Kidd MR, Actor JK, Hunter RL. Evaluation of electrochemiluminescence- and bioluminescence-based assays for quantitating specific DNA. J Clin Lab Anal. 1996; 10:423–31.

17. Hayward-Lester A, Oefner PJ, Sabatini S, Doris PA. Accurate and absolute quantitative measurement of gene expression by single-tube RT-PCR and HPLC. Genome Res. 1995; 5:494–9.

18. Borson ND, Strausbauch MA, Wettstein PJ, Oda RP, Johnston SL, Landers JP. Direct quantitation of RNA transcripts by competitive single-tube RT-PCR and capillary electrophoresis. Biotechniques. 1998; 25:130–7.

19. Huber R, Kunisch E, Gluck B, Egerer R, Sickinger S, Kinne RW. Comparison of conventional and real-time RT-PCR for the quantitation of jun protooncogene mRNA and analysis of junB mRNA expression in synovial membranes and isolated synovial fibroblasts from rheumatoid arthritis patients. Z Rheumatol. 2003; 62:378–89.
20. Bradford WD, Cahoon L, Freel SR, Hoopes LL, Eckdahl TT. An inexpensive gel electrophoresis-based polymerase chain reaction method for quantifying mRNA levels. Cell Biol Educ. 2005; 4:157–68.

21. Frade JP, Warnock DW, Arthington-Skaggs BA. Rapid quantification of drug resistance gene expression in Candida albicans by reverse transcriptase LightCycler PCR and fluorescent probe hybridization. J Clin Microbiol. 2004; 42:2085–93.
Fig. 1.
(A) Generation of an external standard curve for real-time PCR (a: 7 × 10-6, b: 7 × 10-7, c: 7 × 10-8, d: 7 × 10-9 ng/μL). The graph shows the interrelationship between log F (fluorescence) and the amplification cycle; background levels are indicated by the horizontal line. The cycle at which the sample fluorescence crosses the background line is set as the threshold cycle (CT). (B) Standard curve of MMP-1 for real-time PCR. The graph shows the regression curve resulting from threshold determinations for the serially diluted MMP-1 PCR products.
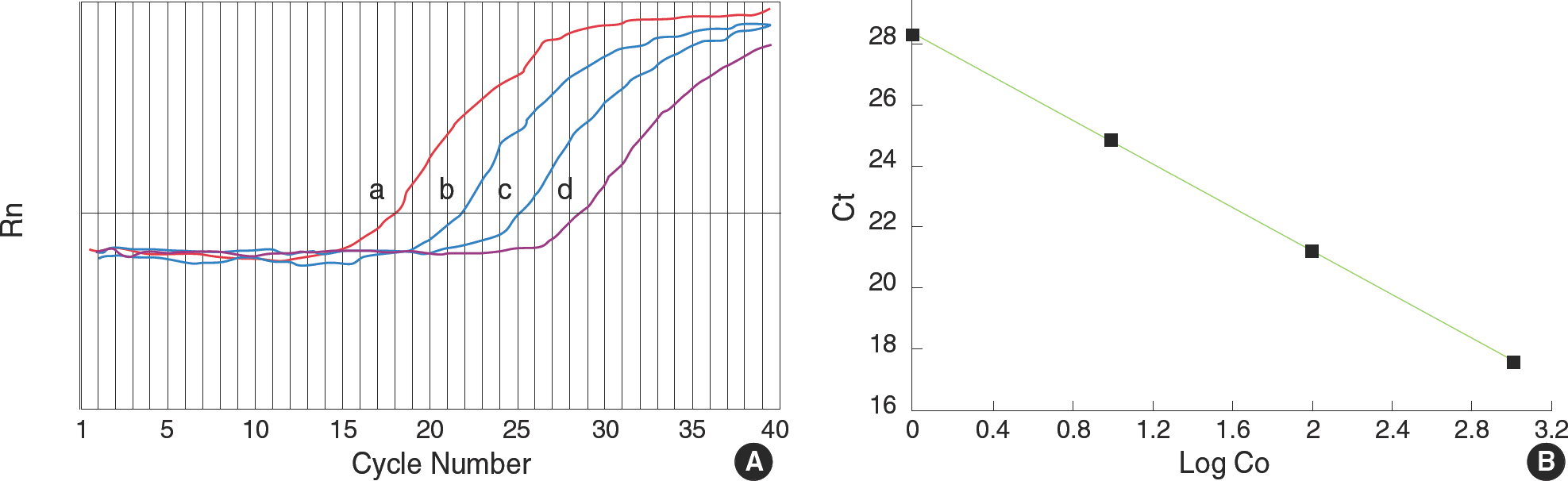
Fig. 2.
Quantification of MMP-1 mRNA by RT-PCR and ethidium bromide-stained agarose gel electrophoresis. Quantification of MMP-1 and GAPDH was accomplished using densitometry of the dye binding intensity of UV-irradiated PCR products. A standard curve was calculated with four values (a, b, c, d) ranging from 7 × 10-6 to 7 × 10-9 ng/L (MMP-1) or 9 × 10-6 to 9 × 10-9 ng/L (GAPDH) PCR product and serves for the quantification of other samples. Expression of MMP-1 was normalized to that of GAPDH expression in the same sample.

Fig. 3.
Comparison between real-time PCR and end-point PCR in (A) MMP-1 mRNA (B) GAPDH mRNA (C) relative quantification of MMP-1 (MMP-1 mRNA/GAPDH mRNA).
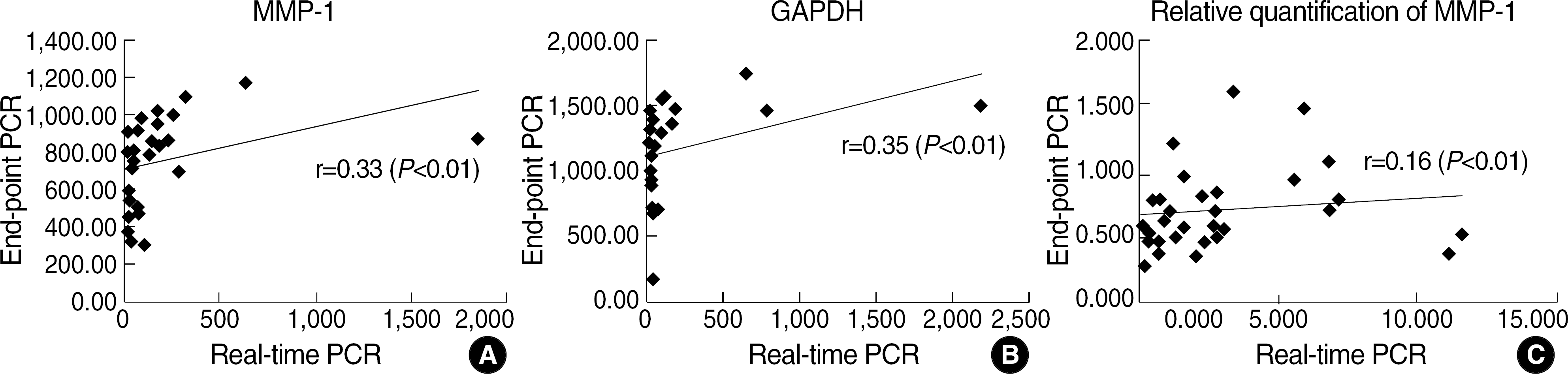
Table 1.
Primers used for PCR
|
Primers |
Oligonucleotide sequence (5′-3′) | Product size (bp) | |
|---|---|---|---|
| MMP-1 | Forward | TGGGATTTCCAAAAGAGGTG | 121 |
| Reverse | ACGTGGTTCCCTGAGAAGA | ||
| GAPDH | Forward | AATGTATCCGTTGTGGATCTGA | 122 |
| Reverse | AGCCCAGGATGCCCTTTA | ||
Table 2.
Comparison between real-time PCR and end-point PCR for relative quantification of MMP-1 mRNA




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download