Abstract
Background
The diagnosis of diseases caused by nontuberculous mycobacteria (NTM) is difficult, because NTM are prevalent in the environment such as soil and water, and because they have fastidious properties. In this study we investigated clinical isolates of NTM for their distribution pattern and accurate species identification.
Methods
We selected presumptive NTM isolates negative for probe hybridization for M. tuberculosis complex, cultured in a third referral hospital from 21 January 2003 to 20 January 2004. Ninety seven-isolates were identified to the species level by direct sequencing of fragments of 16S rRNA, hsp65 and rpoB genes. A total of 120 isolates were studied for the distribution analysis.
Results
Frequently identified NTM species were M. avium (30.8%), M. intracellulare (23.3%) and M. abscessus (18.3%). Others were M. gordonae, M. senegalense, M. fortuitum, M. peregrinum, M. kansasii, M. terrae complex, M. lentiflavum, M. chelonae, and M. szulgai. Three M. tuberculosis complex (2.5%) were also identified among the presumptive NTM isolates. The identification rate by sequencing of 16S rRNA, rpoB, and hsp65 were 65%, 82% and 87%, respectively. The hsp65 or rpoB gene was more efficient than 16S rRNA for the identification of NTM by sequencing.
Go to : 
References
1. Tenholder MF, Moser RJ 3rd, Tellis CJ. Mycobacteria other than tuberculosis. Pulmonary involvement in patients with acquired immunodeficiency syndrome. Arch Intern Med. 1988; 148:953–5.

2. Falkinham JO 3rd. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996; 9:177–215.

3. Debrunner M, Salfinger M, Brandli O, von Graevenitz A. Epidemiology and clinical significance of nontuberculous mycobacteria in patients negative for human immunodeficiency virus in Switzerland. Clin Infect Dis. 1992; 15:330–45.

4. Dobos KM, Quinn FD, Ashford DA, Horsburgh CR, King CH. Emergence of a unique group of necrotizing mycobacterial diseases. Emerg Infect Dis. 1999; 5:367–78.

5. Collins MT, Lisby G, Moser C, Chicks D, Christensen S, Reichelderfer M, et al. Results of multiple diagnostic tests for Mycobacterium avium subsp. paratuberculosis in patients with inflammatory bowel disease and in controls. J Clin Microbiol. 2000; 38:4373–81.
6. Kim SJ, Hong YP, Kim SC, Bai GH, Jin BW, Park CD. A case of pulmonary disease due to M. avium-intracellulare complex. Tuberc Respir Dis. 1981; 28:121–4.
7. Bai GH, Park KS, Kim SJ. Clinically isolated mycobacteria other than mycobacterium tuberculosis from 1980 to 1990 in Korea. J Korean Soc Microbiol. 1993; 28:1–6.
8. American Thoracic Society. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am J Respir Crit Care Med. 1997; 156:S1–25.
9. Pae HH, Lee JH, Yoo CG, Lee CT, Chung HS, Kim YW, et al. Study for clinical characteristics of nontuberculous mycobacterial pulmonary disease. Tuberc Respir Dis. 1999; 47:735–46.
10. Lew WJ, Ahn DI, Yoon YJ, Cho JS, Kwon DW, Kim SJ, et al. Clinical experience on mycobacterial disease other than tuberculosis. Tuberc Respir Dis. 1992; 39:425–32.
11. Lee HW, Kim MN, Shim TS, Bai GH, Pai CH. Nontuberculous mycobacterial pulmonary infection in immunocompetent patients. Tuberc Respir Dis. 2002; 53:173–82.

12. Koh WJ, Kwon OJ, Kang EH, Jeon IS, Pyun YJ, Ham HS, et al. Clinical and radiographic characteristics of 12 patients with Mycobacterium abscessus pulmonary disease. Tuberc Respir Dis. 2003; 54:45–56.
13. Springer B, Stockman L, Teschner K, Roberts GD, Bottger EC. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996; 34:296–303.

14. Brown-Elliott BA, Griffith DE, Wallace RJ Jr. Newly described or emerging human species of nontuberculous mycobacteria. Infect Dis Clin North Am. 2002; 16:187–220.

15. Patel JB, Leonard DG, Pan X, Musser JM, Berman RE, Nachamkin I. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000; 38:246–51.
16. Kim BJ, Lee SH, Lyu MA, Kim SJ, Bai GH, Chae GT, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). J Clin Microbiol. 1999; 37:1714–20.
17. Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001; 39:3637–48.
18. Ringuet H, Akoua-Koffi C, Honore S, Varnerot A, Vincent V, Berche P, et al. hsp65 sequencing for identification of rapidly growing mycobacteria. J Clin Microbiol. 1999; 37:852–7.
19. Harmsen D, Rothganger J, Frosch M, Albert J. RIDOM: Ribosomal Differentiation of Medical Micro-organisms Database. Nucleic Acids Res. 2002; 30:416–7.

20. Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993; 31:175–8.

21. Pai S, Esen N, Pan X, Musser JM. Routine rapid Mycobacterium species assignment based on species-specific allelic variation in the 65-kilodalton heat shock protein gene (hsp65). Arch Pathol Lab Med. 1997; 121:859–64.
22. Kim BJ, Lee KH, Park BN, Kim SJ, Bai GH, Kim SJ, et al. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). J Clin Microbiol. 2001; 39:2102–9.
23. Adekambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003; 41:5699–708.
24. Koh WJ, Kwon OJ, Yu CM, Jeon K, Suh GY, Chung MP, et al. Recovery rate of nontuberculous mycobacteria from acid-fast-bacilli smear-positive sputum specimen. Tuberc Respir Dis. 2003; 54:22–32.
25. Good RC, Snider DE Jr. Isolation of nontuberculous mycobacteria in the United States, 1980. J Infect Dis. 1982; 146:829–33.

26. O'Brien RJ, Geiter LJ, Snider DE Jr. The epidemiology of nontuberculous mycobacterial disease in the United States. Results from a national survey. Am Rev Respir Dis. 1987; 135:1007–14.
27. Ostroff S, Hutwagner L, Collin S. Mycobacterial species and drug resistance patterns reported by state laboratories-1992. 93rd American Society for Microbiology General Meeting. May 16, 1993. Atlanta, GA. Abstract U-9, P.170.
28. Scientific committee in Korean academy of tuberculosis and respiratory disease. National survey of mycobacterial disease other than tuberculosis in Korea. Tuberc Respir Dis. 1995; 42:277–94.
Go to : 
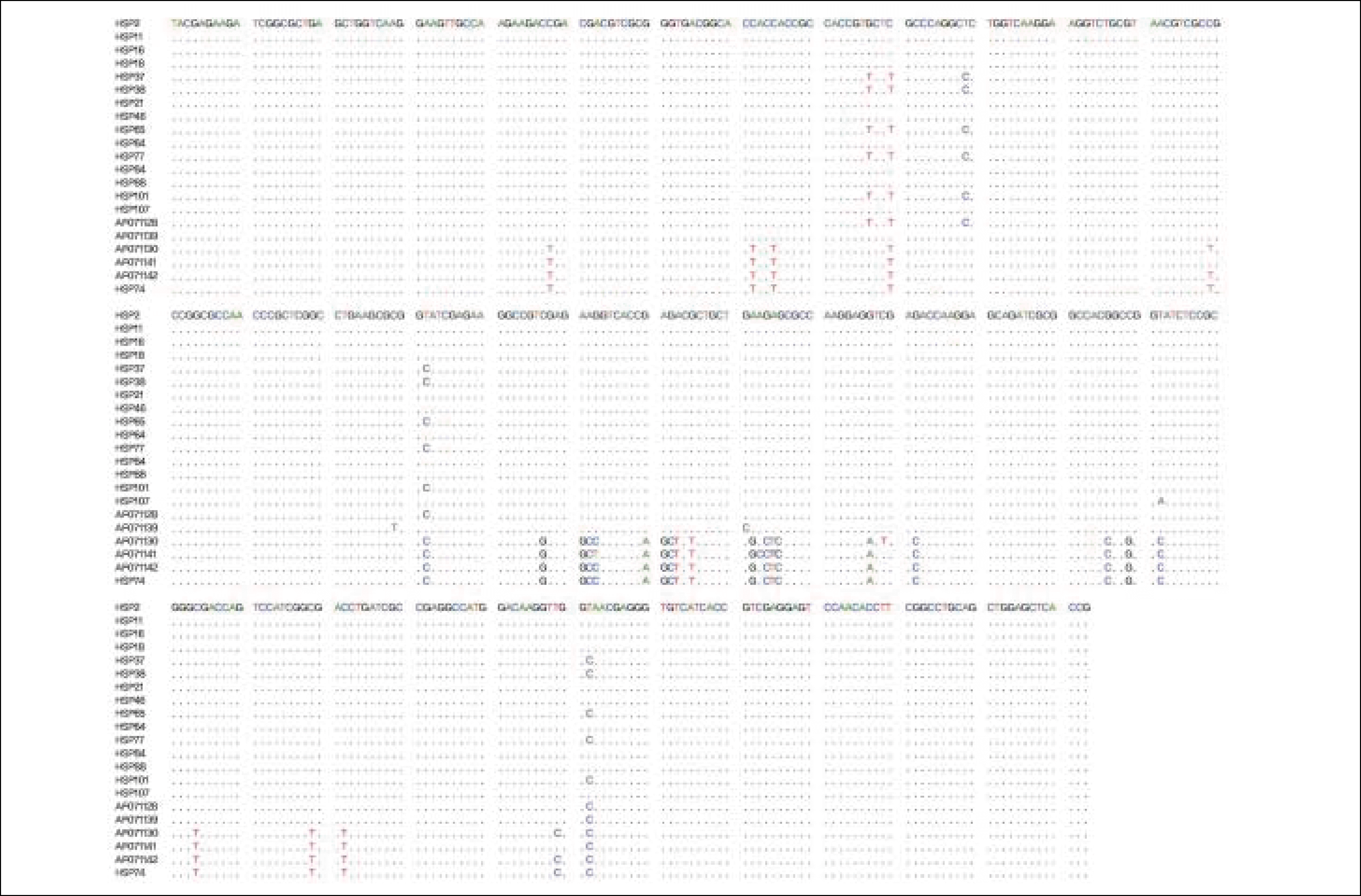 | Fig. 1.Nucleotide sequences of the partial hsp65 gene from 15 M. abscessus(HSP2-HSP107) and one M. chelonae (HSP74) strains aligned with reference sequences from Genebank (AF071128 and AF071139, M. abscessus strains; AF071130, AF071141 and AF 071142, M. chelonae strains). |
Table 1.
Primers for amplification of 16S rRNA, rpoB and hsp65 genes
| Genes | Primers (5′ → 3′) | Amplicon | Ta* (°C) | Reference |
|---|---|---|---|---|
| 16S rRNA | F:AGT TTG ATC CTG GCT CAG | 527 bp | 53 | 17 |
| R:GTA TTG CCG CGG CTG CTG | 19 | |||
| rpoB | F:CGA CCA CTT CGG CAA CCG | 351 bp | 60 | 16 |
| R:TCG ATC GGG CAC ATC CGG | ||||
| hsp65 | F:ACC AAC GAT GGT GTG TCC AT | 441 bp | 53 | 20 |
| R:CTT GTC GAA CCG CAT ACC CT |
Table 2.
Similarity search results using 16S rRNA gene sequence
| First rank | N | Overlap (bp) | Identity (%) | Second rank | Inter-species difference (bp) |
|---|---|---|---|---|---|
| Identified: 62 isolates | |||||
| M. avium | 26 | 415–434 | 99.0–100 | M. lepraemurium | 1 |
| M. fortuitum | 3 | 352–404 | 99.4–100 | M. mucogenicum | 1 |
| M. gordonae | 3 | 419–424 | 100 | M. kansasii | 15 |
| M. intracellulare | 24 | 417–431 | 99.3–100 | M. avium | 2–9 |
| M. lentiflavum | 1 | 411 | 100 | M. simiae | 5 |
| M. szulgai | 1 | 420 | 100 | M. malmoense | 5 |
| M. terrae complex | 2 | 399–423 | 99.3–99.7 | M. hiberniae | 5–7 |
| MTBC | 2 | ||||
| Unidentified: 33 isolates | |||||
| M. abscessus* | 14 | 401–420 | 99.0–100 | M. fortuitum | 8–11 |
| M. kansasii† | 2 | 416–420 | 99.8–100 | M. malmoense | 10 |
| M. peregrinum‡ | 3 | 407–423 | 99.8–100 | M. alvei | 2 |
| M. senegalense§ | 3 | 404–406 | 100 | M. mucogenicum | 1 |
| Bacillus sp. | 3 | ||||
| Signal unsatisfied | 8 | ||||
Table 3.
Final identification of the isolates difficult to identify with a single gene fragment
| Final identification/Genes | 16S rDNA | rpoB | hsp65 |
|---|---|---|---|
| M. avium | Bacillus sp | M. avium | NA† |
| M. abscessus | Bacillus sp | M. abscessus | M. abscessus |
| M. avium | Bacillus sp | M. avium | M. avium |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | M. abscessus | NA† | M. abscessus |
| M. abscessus | MS* | NA† | M. abscessus |
| unidentifiable | MS* | M. gordonae‡ | NA† |
| M. gordonae | MS* | M. gordonae | M. gordonae |
| M. avium | MS* | M. avium | M. avium |
| M. avium | MS* | M. avium | M. avium |
| M. fortuitum | MS* | M. fortuitum | M. fortuitum |
| M. fortuitum | MS* | M. fortuitum | M. fortuitum |
| M. peregrinum | MS* | M. peregrinum | M. peregrinum |
| M. gordonae | M. gordonae | NA† | M. gordonae |
| M. gordonae | M. gordonae | NA† | M. gordonae |
| M. intracellulare | M. intracellulare | NA† | MS* |
| M. kansasii | M. kansasii | NA† | NA† |
| M. avium | M. avium | M. avium | NA† |
| M. avium | M. avium | M. avium | MS* |
Table 4.
Similarity search results using rpoB sequence
Table 5.
Similarity search results using hsp65 sequence
| First rank | N | Overlap (bp) | Identity (%) | Second rank | Inter-species difference (bp) |
|---|---|---|---|---|---|
| Identified: 83 isolates | |||||
| M. abscessus | 15 | 374–396 | 99.2–100 | M. chelonae | 25–28 |
| M. avium | 27 | 352–406 | 99.7–100 | M. intracellulare | 3–10 |
| M. chelonae | 1 | 396 | 99.7 | M. abscessus | 28 |
| M. fortuitum | 5 | 381–392 | 100 | M. senegalense | 3 |
| M. gordonae | 4 | 389–403 | 97.5–99.5 | M. asiaticum | 8–16 |
| M. intracellulare | 22 | 380–406 | 99.2–100 | M. avium | 7–16 |
| M. kansasii | 1 | 386 | 100 | M. gastri | 8 |
| M. lentiflavum | 1 | 382 | 100 | M. triplex | 9 |
| M. peregrinum | 4 | 374–381 | 100 | M. septicum | 3 |
| M. szulgai | 1 | 383 | 100 | M. lentiflavum | 14 |
| MTB complex | 2 | ||||
| Unidentified: 12 isolates | |||||
| M. senegalense* | 3 | 374–396 | 98.2–98.5 | M. fortuitum | 1 |
| M. avium | 1 | 396 | 98.0 | M. intracellulare | 3 |
| M. terrae complex | 2 | 398 | 97.0 | M. abscessus | 8–16 |
| No amplicon | 4 | ||||
| Signal unsatisfied | 2 |
Table 6.
Distribution of MTBC probe negative, presumptive NTM isolates
| Organism | No. case | Percent (%) |
|---|---|---|
| M. avium | 37 | 30.8 |
| M. intracellulare | 28 | 23.3 |
| M. abscessus | 22 | 18.3 |
| M. fortuitum | 8 | 6.7 |
| M. peregrinum | 4 | 3.3 |
| M. gordonae | 3 | 2.5 |
| M. senegalense | 3 | 2.5 |
| M. kansasii | 2 | 1.7 |
| M. terrae complex | 3 | 1.7 |
| M. chelonae | 1 | 0.8 |
| M. celatum | 1 | 0.8 |
| M. lentiflavum | 1 | 0.8 |
| M. szulgai | 1 | 0.8 |
| MTB complex | 3 | 2.5 |
| Oral contaminant | 2 | 1.7 |
| unidentifiable* | 1 | 0.8 |
| Total | 120 | 100 |
Table 7.
Comparison of the three gene segments used in identification of presumptive NTM isolates
| Genes | Cases* | Presence of amplicon (%) | Identification of NTM (%) |
|---|---|---|---|
| 16S rRNA | 95 | 100 | 65 |
| rpoB | 95 | 87 | 82 |
| hsp65 | 95 | 96 | 71† |
| 87‡ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download