Abstract
Colorectal cancer (CRC) is one of the leading causes of cancer related deaths in the world. Many oncogenes and tumor suppressor genes are involved in the development of CRC. MicroRNAs (miRNAs) are a class of small, non-coding, endogenous RNAs in animals and plants. Recent studies have shown that miRNAs are associated with the mediation process of tumorigenesis, including inflammation, cell cycle, stress response, differentiation, apoptosis, migration, and invasion in cancer. These miRNAs have been linked to the development of CRC and recently studied as new potential biomarkers in the diagnosis and treatment for CRC. Specific miRNAs expression patterns help distinguish CRC from other colon-related diseases, and miRNAs can target the oncogenes and regulatory molecular pathways. Recent studies have demonstrated the restoration of tumor suppressive miRNAs and inhibition of oncogenic miRNAs for CRC treatment. Herein, we describe the diagnostic and therapeutic roles of miRNAs in CRC.
Go to : 
References
1. Meyerhardt JA, Giovannucci EL, Holmes MD, et al. Physical activity and survival after colorectal cancer diagnosis. J Clin Oncol. 2006; 24:3527–3534.

2. Hegewisch-Becker S, Graeven U, Arnold D. Maintenance treatment in metastatic colorectal cancer – authors'reply. Lancet Oncol. 2015; 16:e584–e585.
3. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854.

4. Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature. 2000; 403:901–906.

7. Lai EC. Predicting and validating microRNA targets. Genome Biol. 2004; 5:115.
9. Yang Y, Yang JJ, Tao H, Jin WS. MicroRNA-21 controls hTERT via PTEN in human colorectal cancer cell proliferation. J Physiol Biochem. 2015; 71:59–68.

10. Zhang G, Zhou H, Xiao H, Liu Z, Tian H, Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014; 59:98–107.

11. Wu W, Yang J, Feng X, et al. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013; 12:30.

12. Geng L, Sun B, Gao B, et al. MicroRNA-103 promotes colorectal cancer by targeting tumor suppressor DICER and PTEN. Int J Mol Sci. 2014; 15:8458–8472.

13. Ji D, Chen Z, Li M, et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014; 13:86.

14. Wei Z, Cui L, Mei Z, et al. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014; 588:1773–1779.

15. Fang Y, Sun B, Li Z, Chen Z, Xiang J. miR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2016; 55:1369–1377.

16. Wang YX, Chen YR, Liu SS, et al. miR-384 inhibits human colorectal cancer metastasis by targeting KRAS and CDC42. Oncotarget. 2016; 7:84826–84838.

17. Zhou Y, Feng X, Liu YL, et al. Downregulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013; 8:e81203.

18. Zheng K, Liu W, Liu Y, Jiang C, Qian Q. MicroRNA-133a suppresses colorectal cancer cell invasion by targeting Fascin1. Oncol Lett. 2015; 9:869–874.

19. Duan FT, Qian F, Fang K, Lin KY, Wang WT, Chen YQ. miR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol Cancer. 2013; 12:164.

20. Strillacci A, Valerii MC, Sansone P, et al. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013; 229:379–389.
21. Ji S, Ye G, Zhang J, et al. miR-574–5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013; 62:716–726.
22. Li T, Lai Q, Wang S, et al. MicroRNA-224 sustains Wnt/β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016; 35:21.

23. Zhang JX, Mai SJ, Huang XX, et al. miR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann Oncol. 2014; 25:2196–2204.

24. Sun Y, Shen S, Liu X, et al. miR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting one-cut2 in colorectal carcinoma. Mol Cell Biochem. 2014; 390:19–30.

25. Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. Snail and miR-34a feedforward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013; 32:3079–3095.

26. Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014; 124:1853–1867.

27. Park YR, Lee ST, Kim SL, et al. MicroRNA-9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer. Int J Oncol. 2016; 48:2135–2143.

28. Zhu M, Xu Y, Ge M, Gui Z, Yan F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015; 106:833–839.
29. Zhao S, Sun H, Jiang W, et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFbeta-mediated epithelial to mesenchymal transition. Mol Cancer. 2017; 16:12.

30. Xu T, Jing C, Shi Y, et al. microRNA-20a enhances the epithelial-to-mesenchymal transition of colorectal cancer cells by modulating matrix metalloproteinases. Exp Ther Med. 2015; 10:683–688.

31. Fantini S, Salsi V, Reggiani L, Maiorana A, Zappavigna V. The miR-196b miRNA inhibits the GATA6 intestinal transcription factor and is upregulated in colon cancer patients. Oncotarget. 2017; 8:4747–4759.

32. Chen J, Wang W, Zhang Y, Hu T, Chen Y. The roles of miR-200c in colon cancer and associated molecular mechanisms. Tumour Biol. 2014; 35:6475–6483.

33. Kim BK, Yoo HI, Lee AR, Choi K, Yoon SK. Decreased expression of VLDLR is inversely correlated with miR-200c in human colorectal cancer. Mol Carcinog. 2017 Jan 23. [Epub ahead of print].
34. Zhang N, Lu C, Chen L. miR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncol Lett. 2016; 12:4589–4597.

35. Qiu G, Zhang XB, Zhang SQ, et al. Dysregulation of MALAT1 and miR-619–5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016; 12:5036–5042.

36. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a–5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017; 16:9.

37. Li B, Xie Z, Li Z, Chen S, Li B. MicroRNA-613 targets FMNL2 and suppresses progression of colorectal cancer. Am J Transl Res. 2016; 8:5475–5484. eCollection 2016.
38. Fujino Y, Takeishi S, Nishida K, et al. Downregulation of microRNA-100/microRNA-125b is associated with lymph node metastasis in early colorectal cancer with submucosal invasion. Cancer Sci. 2017; 108:390–397.

39. Hata T, Mokutani Y, Takahashi H, et al. Identification of microRNA-487b as a negative regulator of liver metastasis by regulation of KRAS in colorectal cancer. Int J Oncol. 2017; 50:487–496.

40. Zu C, Liu T, Zhang G. MicroRNA-506 Inhibits malignancy of colorectal carcinoma cells by targeting LAMC1. Ann Clin Lab Sci. 2016; 46:666–674.
41. Hao H, Xia G, Wang C, Zhong F, Liu L, Zhang D. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med Mol Morphol. 2016 Dec 15. [Epub ahead of print].

42. Lu W, Wang J, Yang G, et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget. 2017 Jan 27. [Epub ahead of print].

43. Yuan L, Yuan P, Yuan H, et al. miR-542–3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1. Am J Cancer Res. 2017; 7:159–172.
44. Bai JW, Xue HZ, Zhang C. Downregulation of microRNA-143 is associated with colorectal cancer progression. Eur Rev Med Pharmacol Sci. 2016; 20:4682–4687.
45. Yu L, Lu Y, Han X, et al. microRNA-140–5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem Cell Res Ther. 2016; 7:180.

46. Zhang X, Xu J, Jiang T, Liu G, Wang D, Lu Y. MicroRNA-195 suppresses colorectal cancer cells proliferation via targeting FGF2 and regulating Wnt/β-catenin pathway. Am J Cancer Res. 2016; 6:2631–2640. eCollection 2016.
47. Guo F, Luo Y, Mu YF, et al. miR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancer. Am J Cancer Res. 2016; 6:2463–2475. eCollection 2016.
48. Di Lena M, Travaglio E, Altomare DF. New strategies for colorectal cancer screening. World J Gastroenterol. 2013; 19:1855–1860.
49. Yau TO, Wu CW, Dong Y, et al. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer. 2014; 111:1765–1771.

50. Phua LC, Chue XP, Koh PK, Cheah PY, Chan EC, Ho HK. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol Rep. 2014; 32:97–104.

51. Wu CW, Ng SS, Dong YJ, et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012; 61:739–745.

52. Koga Y, Yamazaki N, Yamamoto Y, et al. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013; 22:1844–1852.

53. Wu CW, Ng SC, Dong Y, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014; 20:2994–3002.

54. Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011; 46:1391–1402.

55. Li JM, Zhao RH, Li ST, et al. Downregulation of fecal miR-143 and miR-145 as potential markers for colorectal cancer. Saudi Med J. 2012; 33:24–29.
56. Ghanbari R, Mosakhani N, Asadi J, et al. Decreased expression of fecal miR-4478 and miR-1295b–3p in early-stage colorectal cancer. Cancer Biomark. 2015; 15:189–195.

57. Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010; 25:1674–1680.

58. Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013; 105:849–859.

59. Basati G, Emami Razavi A, Abdi S, Mirzaei A. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med Oncol. 2014; 31:205.

60. Yuan D, Li K, Zhu K, Yan R, Dang C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol Ther. 2015; 16:268–275.

61. Zhang L, Meng L, Fan Z, Liu B, Pei Y, Zhao Z. Expression of plasma miR-106a in colorectal cancer and its clinical significance. Nan Fang Yi Ke Da Xue Xue Bao. 2014; 34:354–357.
62. Chen Q, Xia HW, Ge XJ, Zhang YC, Tang QL, Bi F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac J Cancer Prev. 2013; 14:7421–7426.

63. Brunet Vega A, Pericay C, Moya I, et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep. 2013; 30:320–326.
64. Yamazaki N, Koga Y, Taniguchi H, et al. High expression of miR-181c as a predictive marker of recurrence in stage II colorectal cancer. Oncotarget. 2017; 8:6970–6983.

65. Meng WJ, Yang L, Ma Q, et al. MicroRNA expression profile reveals miR-17–92 and miR-143–145 cluster in synchronous colorectal cancer. Medicine (Baltimore). 2015; 94:e1297.

66. Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010; 12:433–440.

67. Ahmed FE, Ahmed NC, Vos PW, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013; 10:93–113.
68. Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013; 8:e62880.

69. Zheng G, Du L, Yang X, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014; 111:1985–1992.

70. Chu D, Zheng J, Li J, et al. MicroRNA-630 is a prognostic marker for patients with colorectal cancer. Tumour Biol. 2014; 35:9787–9792.

71. Liu Y, Zhou Y, Feng X, et al. Low expression of microRNA-126 is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2014; 53:358–365.

72. Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014; 14:109.

73. Diaz T, Tejero R, Moreno I, et al. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol. 2014; 109:676–683.

74. Zhang Y, Geng L, Talmon G, Wang J. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J Biol Chem. 2015; 290:6215–6225.

75. Zhang Y, Zheng L, Huang J, et al. miR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014; 9:e93917.

Go to : 
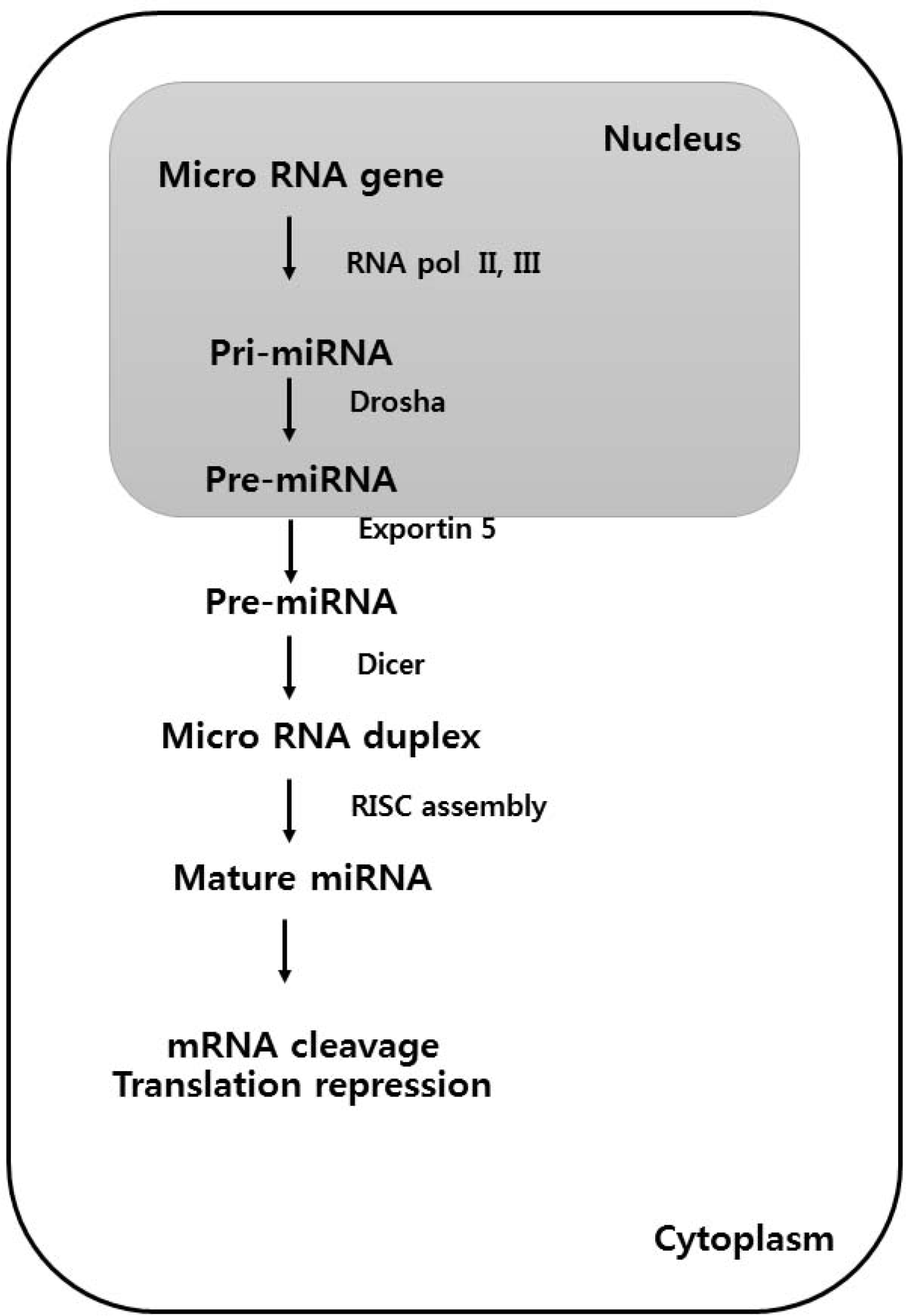 | Fig. 1.Biogenesis of microRNAs. RNA polymerase II (Pol II) produces a primary microRNA (miRNA) that is then cropped to form a pre-miRNA hairpin by a Drosha. This double-stranded hairpin structure is exported from the nucleus by exportin 5. Finally, the pre-miRNA is cleaved by DICER1 a mature miRNA. The single stranded miRNA is incorporated into RISC, which then targets it to the target 3′ untranslated region messenger RNA (mRNA) sequence to facilitate repression and cleavage.5,6
|
Table 1.
Oncogenic or Tumor Suppressor MicroRNAs in Colorectal Cancer
Table 2.
MicroRNAs as Potential Biomarker in Various Samples of Colorectal Cancer




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download