Abstract
Background/Aims
Helicobacter pylori infection induces aberrant DNA methylation in gastric mucosa. We evaluated the long-term effect of H. pylori eradication on promotor CpG island hypermethylation in gastric carcinogenesis.
Methods
H. pylori-positive patients with gastric adenoma or early gastric cancer who underwent endoscopic resection were enrolled. According to H. pylori eradication after endoscopic resection, the participants were randomly assigned to H. pylori eradication or non-eradication group. H. pylori-negative gastric mucosa from normal participants provided the normal control. CpG island hypermethylation of tumor-related genes (p16, CDH1, and RUNX-3) was evaluated by quantitative MethyLight assay in non-tumorous gastric mucosa. The gene methylation rate and median values of hypermethylation were compared after one year by H. pylori status.
Results
In H. pylori-positive patients, hypermethylation of p16 was found in 80.6%, of CDH1 in 80.6%, and of RUNX-3 in 48.4%. This is significantly higher than normal control (p16, 10%; CDH1, 44%; RUNX-3, 16%) (p<0.05). In the H. pylori eradication group, methylation rates of p16 and CDH1 decreased in 58.1% and 61.3% of the patients, and the median values of hyper-methylation were significantly lower at one year compared with the non-eradication group. However, RUNX-3 hypermethylation did not differ significantly at one year after H. pylori eradication. The non-eradication group hypermethylation did not change after one year.
Go to : 
References
1. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994; 61:1–241.
2. Na HK, Woo JH. Helicobacter pylori induces hypermethylation of CpG islands through upregulation of DNA methyltransferase: possible involvement of reactive oxygen/nitrogen species. J Cancer Prev. 2014; 19:259–264.
5. Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001; 61:2847–2851.
6. Maekita T, Nakazawa K, Mihara M, et al. High levels of aberrant DNA methylation in Helicobacter pyloriinfected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006; 12:989–995.
7. Ito K, Liu Q, Salto-Tellez M, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005; 65:7743–7750.

8. Perri F, Cotugno R, Piepoli A, et al. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol. 2007; 102:1361–1371.
9. Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009; 40:1534–1542.

10. Choi J, Kim SG, Yoon H, et al. Eradication of Helicobacter pylori after endoscopic resection of gastric tumors does not reduce incidence of metachronous gastric carcinoma. Clin Gastroenterol Hepatol. 2014; 12:793–800.e1.
11. Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996; 20:1161–1181.
12. Shin CM, Kim N, Jung Y, et al. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci. 2010; 101:1337–1346.
13. Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009; 219:410–416.
14. Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000; 28:E32.

15. Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006; 8:209–217.

16. Shin SH, Park SY, Ko JS, Kim N, Kang GH. Aberrant CpG island hypermethylation in pediatric gastric mucosa in association with Helicobacter pylori infection. Arch Pathol Lab Med. 2011; 135:759–765.
17. Kim JH, Rhee YY, Bae JM, et al. Subsets of microsatellite- unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol. 2013; 26:1013–1022.
18. Chan AO, Peng JZ, Lam SK, et al. Eradication of Helicobacter pylori infection reverses E-cadherin promoter hypermethylation. Gut. 2006; 55:463–468.
19. Tahara T, Arisawa T, Shibata T, et al. Increased number of methylated CpG islands correlates with Helicobacter pylori infection, histological and serological severity of chronic gastritis. Eur J Gastroenterol Hepatol. 2009; 21:613–619.
20. Bartchewsky W Jr, Martini MR, Squassoni AC, et al. Influence of Helicobacter pylori infection on the expression of MLH1 and MGMT in patients with chronic gastritis and gastric cancer. Eur J Clin Microbiol Infect Dis. 2009; 28:591–597.
21. Sepulveda AR, Yao Y, Yan W, et al. CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology. 2010; 138:1836–1844.
22. Leung WK, Man EP, Yu J, et al. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in non-cancerous stomach. Clin Cancer Res. 2006; 12:3216–3221.
23. Kitajima Y, Ohtaka K, Mitsuno M, et al. Helicobacter pylori infection is an independent risk factor for Runx3 methylation in gastric cancer. Oncol Rep. 2008; 19:197–202.
24. Lu XX, Yu JL, Ying LS, et al. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012; 118:5507–5517.
Go to : 
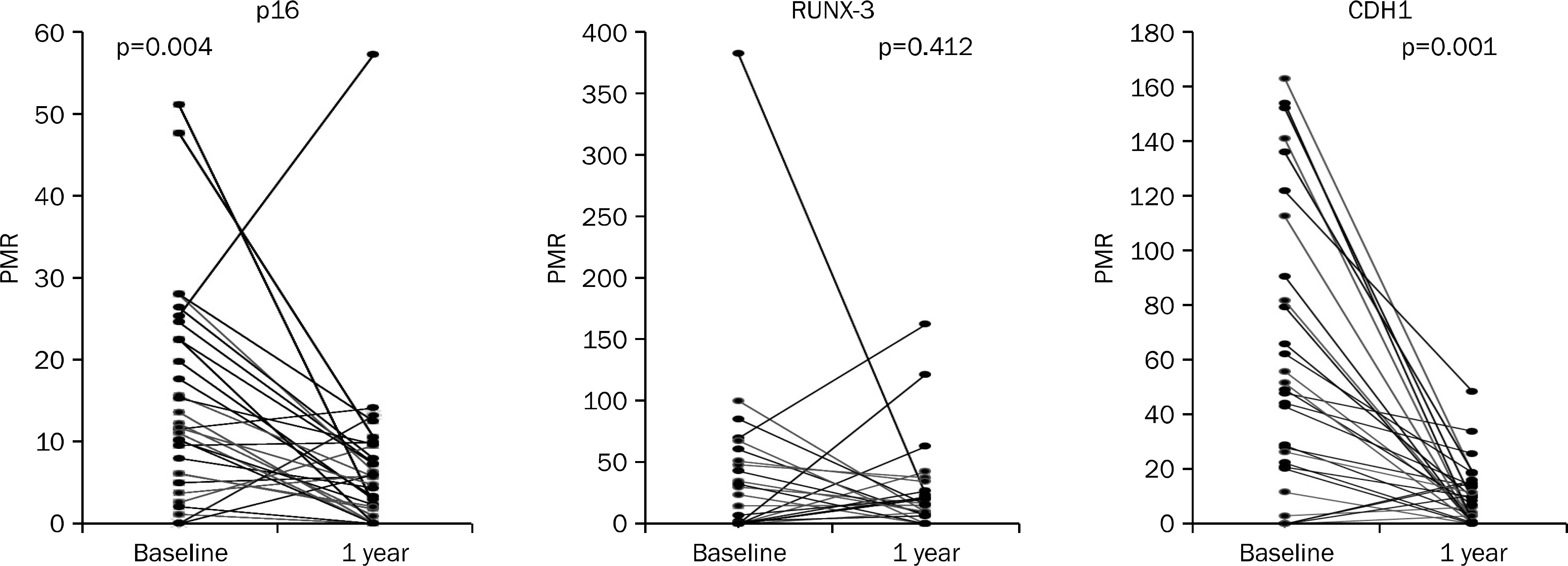 | Fig. 1.Changes in quantitative value of MethyLight assay (percentage of methylated reference, PMR) of p16, RUNX-3 and CDH1 at baseline and after one year in the Helicobacter pylori eradication group. PMR value in p16 and CDH1 were significantly reduced one year after eradication. |
Table 1.
Clinicopathological Characteristics among Groups
| Characteristic | Group | |||
|---|---|---|---|---|
| Helicobacer pylori eradication | Non-eradication | p-value a | Normal control | |
| Gender | 1.00 | |||
| Male | 21 (67.7) | 22 (64.7) | 11 (61.1) | |
| Female | 10 (32.3) | 12 (35.3) | 7 (38.9) | |
| Age (yr) | 60.0 (50.0–66.0) | 59.5 (54.7–66.0) | 0.68 | 58.5 (50.0–67.5) |
| Gastric atrophy | 0.42 | |||
| Absent | 5 (16.1) | 2 (5.9) | 7 (38.9) | |
| Mild | 6 (19.4) | 6 (17.6) | 9 (50.0) | |
| Moderate | 11 (35.5) | 10 (29.4) | 2 (11.1) | |
| Marked | 3 (9.7) | 6 (17.6) | 0 (0) | |
| NA | 6 (19.4) | 10 (29.4) | 0 (0) | |
| Intestinal metaplasia | 1.00 | |||
| Absent | 1 (3.2) | 2 (5.9) | 11 (61.1) | |
| Mild | 9 (29.0) | 8 (23.5) | 3 (16.7) | |
| Moderate | 13 (41.9) | 19 (55.9) | 4 (22.2) | |
| Marked | 8 (25.8) | 5 (14.7) | 0 (0) | |
| Gastric atrophy or IM | 0.97 | |||
| Absent | 1 (3.2) | 1 (2.9) | 7 (38.9) | |
| Mild | 9 (29.0) | 9 (26.5) | 9 (50.0) | |
| Moderate | 12 (38.7) | 15 (44.1) | 2 (11.1) | |
| Marked | 9 (29.0) | 9 (26.5) | 0 (0) | |
| H. pylori density | 0.75 | 0 (0) | ||
| Mild | 10 (32.3) | 14 (41.2) | ||
| Moderate | 14 (45.2) | 13 (38.2) | ||
| Marked | 7 (22.6) | 7 (20.6) | ||
| Neutrophilic inflammation | 0.25 | |||
| Absent | 1 (3.2) | 0 (0) | ||
| Mild | 0 (0) | 3 (8.8) | ||
| Moderate | 25 (80.6) | 27 (79.4) | ||
| Marked | 5 (16.1) | 4 (11.8) | ||
| Mononuclear inflammation | 0.43 | |||
| Absent | 0 (0) | 0 (0) | ||
| Mild | 0 (0) | 0 (0) | ||
| Moderate | 20 (64.5) | 25 (73.5) | ||
| Marked | 11 (35.5) | 9 (26.5) | ||
| Histology | 0.55 | |||
| Low-grade dysplasia | 12 (38.7) | 10 (29.4) | 0 (0) | |
| High-grade dysplasia | 4 (12.9) | 3 (8.8) | 0 (0) | |
| Adenocarcinoma | 15 (48.4) | 21 (61.8) | 0 (0) | |
| Well differentiated | 5 (16.1) | 11 (32.4) | ||
| Moderately differentiated | 7 (22.6) | 6 (17.6) | ||
| Poorly differentiated | 2 (6.5) | 1 (2.9) | ||
| Signet ring cell | 1 (3.2) | 3 (8.8) | ||
| Total | 31 (100) | 34 (100) | 18 (100) | |
Table 2.
Promotor Gene Methylation Rates and Median Methylation Values at Baseline and One Year among Groups
| Variable | Group | |||||||
|---|---|---|---|---|---|---|---|---|
| Helicobacter pylori eradication | Non-eradication | Normal control | ||||||
| Baseline | 1 yr | p-value a | Baseline | 1 yr | p-value b | Baseline | p-value c | |
| Methylated gene (%) | ||||||||
| P16 | 80.6 | 58.1 | 1.00 | 71.1 | 84.2 | 0.33 | 10.0 | 0.01 |
| RUNX-3 | 48.4 | 67.7 | 0.25 | 55.3 | 65.8 | 0.05 | 16.0 | 0.03 |
| CDH1 | 80.6 | 61.3 | 1.00 | 97.4 | 100 | 1.00 | 44.0 | 0.01 |
| Median methylation value (PMR) | ||||||||
| P16 | 11.7 | 5.7 | 0.01 | 12.9 | 17.8 | 0.25 | 3.4 | 0.01 |
| RUNX-3 | 2.2 | 9.8 | 0.41 | 30.5 | 50.4 | 0.18 | 13.9 | 0.01 |
| CDH1 | 47.9 | 7.0 | 0.01 | 67.7 | 76.6 | 0.24 | 5.3 | 0.01 |
| Patient (total, n) | 31 | 34 | 18 | |||||
Table 3.
Promotor Gene Methylation Rates and Median Methylation Values at Baseline and One Year according to Gastric Adenoma or Early Gastric Cancer




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download