Abstract
A complex microbiota colonizes mucosal layers in different regions of the human gut. In the healthy state, the microbial communities provide nutrients and energy to the host via fermentation of non-digestible dietary components in the large intestine. In contrast, they can play roles in inflammation and infection, including gastrointestinal diseases and metabolic syndrome such as obesity. However, because of the complexity of the microbial community, the functional connections between the enteric microbiota and metabolism are less well understood. Nevertheless, major progress has been made in defining dominant bacterial species, community profiles, and systemic characteristics that produce stable microbiota beneficial to health, and in identifying their roles in enteric metabolism. Through studies in both mice and humans, we are recently in a better position to understand what effect the enteric microbiota has on the metabolism by improving energy yield from food and modulating dietary components. Achieving better knowledge of this information may provide insights into new possibilities that reconstitution of enteric microbiota via diet can provide the maintenance of healthy state and therapeutic/preventive strategies against metabolic syndrome such as obesity. This review focuses on enteric microbial composition and metabolism on healthy and diseased states.
Go to : 
References
1. Kang JS. Journal walk regarding the expanding role of microbiota in our gut. J Bacteriol Virol. 2011; 41:63–64.

2. Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008; 6:776–788.

3. Booijink CC, El-aidy S, Rajilić-stojanović M, et al. High temporal and interindividual variation detected in the human ileal microbiota. Environ Microbiol. 2010; 12:3213–3227.

4. Zoetendal EG, Raes J, van den Bogert B, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012; 6:1415–1426.

5. Hartman AL, Lough DM, Barupal DK, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A. 2009; 106:17187–17192.

6. Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005; 54:219–231.

7. Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol. 2009; 11:2112–2122.

8. Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005; 71:3692–3700.

9. Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004; 427:441–444.

10. Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and com-petition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007; 9:1101–1111.

11. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008; 105:13580–13585.

12. Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011; 141:1773–1781.

14. Eggesbø M, Moen B, Peddada S, et al. Development of gut microbiota in infants not exposed to medical interventions. APMIS. 2011; 119:17–35.

15. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010; 107:11971–11975.

16. Karlsson CL, Molin G, Cilio CM, Ahrné S. The pioneer gut microbiota in human neonates vaginally born at term-a pilot study. Pediatr Res. 2011; 70:282–286.

17. Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010; 86(Suppl 1):13–15.

18. Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery – effects on gut microbiota and humoral immunity. Neonatology. 2008; 93:236–240.

19. Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010; 51:77–84.

20. Klaassens ES, Boesten RJ, Haarman M, et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl Environ Microbiol. 2009; 75:2668–2676.

21. Harmsen HJ, Wildeboer-veloo AC, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000; 30:61–67.

22. Roger LC, Mccartney AL. Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiology. 2010; 156:3317–3328.

23. Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007; 14:169–181.

24. Martín R, Jiménez E, Heilig H, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009; 75:965–969.

25. Solís G, de Los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010; 16:307–310.

26. Magne F, Abély M, Boyer F, Morville P, Pochart P, Suau A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006; 57:128–138.

27. Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002; 68:219–226.

28. Fallani M, Amarri S, Uusijarvi A, et al. INFABIO team. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011; 157(Pt 5):1385–1392.

29. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012; 486:222–227.

30. Martin R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010; 1:367–382.

31. Kalliomäki M, Isolauri E. Pandemic of atopic diseases–a lack of microbial exposure in early infancy? Curr Drug Targets Infect Disord. 2002; 2:193–199.
32. Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007; 56:661–667.

33. O'Toole PW, Claesson MJ. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010; 20:281–291.
35. Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4586–4591.

37. Kim HS. Our genome and our other genome: understanding humans as symbionts with microbes. J Bacteriol Virol. 2012; 42:101–107.

38. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012; 489:242–249.

40. Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012; 9:577–589.

41. Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009; 11:2574–2584.

42. Walker AW, Ince J, Duncan SH, et al. Dominant and diet-re-sponsive groups of bacteria within the human colonic microbiota. ISME J. 2011; 5:220–230.

43. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010; 107:14691–14696.

44. Kim W. Application of metagenomic techniques: understanding the unrevealed human microbiota and explaining the in clinical infectious diseases. J Bacteriol Virol. 2012; 42:263–275.

45. Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007; 449:843–849.

46. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334:105–108.

47. Roberfroid M, Gibson GR, Hoyles L, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010; 104(Suppl 2):S1–S63.

48. Davis LM, Martínez I, Walter J, Goin C, Hutkins RW. Barcoded pyrosequencing reveals that consumption of galactooligosa-ccharides results in a highly specific bifidogenic response in humans. PLoS One. 2011; 6:e25200.

49. Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009; 101:541–550.
50. Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006; 312:1355–1359.

51. Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007; 102:1197–1208.

52. Van Wey AS, Cookson AL, Roy NC, McNabb WC, Soboleva TK, Shorten PR. Bacterial biofilms associated with food particles in the human large bowel. Mol Nutr Food Res. 2011; 55:969–978.

53. Leitch EC, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007; 9:667–679.

54. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012; 6:1535–1543.

55. Walker AW, Duncan SH, Harmsen HJ, Holtrop G, Welling GW, Flint HJ. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008; 10:3275–3283.

56. Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex gly-can catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009; 284:24673–24677.

57. Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysacchar-ide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008; 6:121–131.

58. van Passel MW, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin de-grader, and its use in exploring intestinal metagenomes. PLoS One. 2011; 6:e16876.
59. Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011; 2:166.

60. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3:289–306.

61. Sleeth ML, Thompson EL, Ford HE, Zac-Varghese SE, Frost G. Free fatty acid receptor 2 and nutrient sensing: a proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr Res Rev. 2010; 23:135–145.

62. Gassull MA. Review article: the intestinal lumen as a therapeutic target in inflammatory bowel disease. Aliment Pharmacol Ther. 2006; 24(Suppl 3):90–95.

63. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008; 27:104–119.

64. Lewis SJ, Heaton KW. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut. 1997; 41:245–251.

65. Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut. 1994; 35(1 Suppl):S35–S38.

66. Gao Z, Yin J, Zhang J, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009; 58:1509–1517.

67. Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the bu-tyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010; 12:304–314.

68. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009; 294:1–8.

69. Aminov RI, Walker AW, Duncan SH, Harmsen HJ, Welling GW, Flint HJ. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacte-rium rectale. Appl Environ Microbiol. 2006; 72:6371–6376.
70. Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. Cell-associated alpha-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology. 2006; 152(Pt 11):3281–3290.
71. Scott KP, Martin JC, Chassard C, et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4672–4679.

72. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pec-tin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012; 78:420–428.

73. Brinkworth GD, Noakes M, Clifton PM, Bird AR. Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr. 2009; 101:1493–1502.

74. Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007; 73:1073–1078.

75. Russell WR, Gratz SW, Duncan SH, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr. 2011; 93:1062–1072.

76. El Oufir L, Flourié B, Bruley des Varannes S, et al. Relations between transit time, fermentation products, and hydrogen con-suming flora in healthy humans. Gut. 1996; 38:870–877.

77. McOrist AL, Miller RB, Bird AR, et al. Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J Nutr. 2011; 141:883–889.

78. Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004; 70:5810–5817.

79. Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007; 73:6526–6533.

80. Belenguer A, Holtrop G, Duncan SH, et al. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol Ecol. 2011; 77:107–119.

81. Bourriaud C, Robins RJ, Martin L, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but interindividual variation is evident. J Appl Microbiol. 2005; 99:201–212.

82. Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr. 2006; 96:570–577.
83. Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988; 95:1564–1568.

84. Smith EA, Macfarlane GT. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998; 25:355–368.

85. Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003; 62:67–72.

86. Medani M, Collins D, Docherty NG, Baird AW, O'Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis. 2011; 17:1620–1625.

87. Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006; 4:9–14.

88. Kim JM. Inflammatory bowel diseases and enteric microbiota. Korean J Gastroenterol. 2010; 55:4–18.

89. Marquet P, Duncan SH, Chassard C, Bernalier-Donadille A, Flint HJ. Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett. 2009; 299:128–134.

90. Sahakian AB, Jee SR, Pimentel M. Methane and the gastrointestinal tract. Dig Dis Sci. 2010; 55:2135–2143.

91. Rey FE, Faith JJ, Bain J, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010; 285:22082–22090.

92. Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydro-genotrophic microbes in the healthy human colon. ISME J. 2012; 6:57–70.

93. Possemiers S, Bolca S, Verstraete W, Heyerick A. The intestinal microbiome: a separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia. 2011; 82:53–66.

94. McIntosh FM, Maison N, Holtrop G, et al. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ Microbiol. 2012; 14:1876–1887.

95. Gloux K, Berteau O, El Oumami H, Béguet F, Leclerc M, Doré J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4539–4546.

96. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009; 106:3698–3703.

97. Na HN, Nam JH. Infectobesity: a new area for microbiological and virological research. J Bacteriol Virol. 2011; 41:65–76.

98. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–1031.

100. Roberfroid MB. Caloric value of inulin and oligofructose. J Nutr. 1999; 129(7 Suppl):1436S–1437S.

101. Parnell JA, Reimer RA. Prebiotic fibres dose-dependently increase satiety hormones and alter Bacteroidetes and Firmi-cutes in lean and obese JCR:LA-cp rats. Br J Nutr. 2012; 107:601–613.

102. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006; 444:1022–1023.
103. Schwiertz A, Taras D, Schäfer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010; 18:190–195.

104. Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond). 2008; 32:1720–1724.

105. Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004; 101:15718–15723.
106. Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009; 137:1716–1724.

107. Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010; 59:1635–1642.

108. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009; 1:6ra14.

109. Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010; 328:228–231.

110. Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010; 104:919–929.

111. Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia ini-tiates obesity and insulin resistance. Diabetes. 2007; 56:1761–1772.
112. Stephen AM, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut. 1987; 28:601–609.

113. Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Curr Opin Clin Nutr Metab Care. 2007; 10:729–734.

114. Caesar R, Reigstad CS, Bäckhed HK, et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012; 61:1701–1707.

115. Creely SJ, McTernan PG, Kusminski CM, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007; 292:E740–E747.

116. Amar J, Burcelin R, Ruidavets JB, et al. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008; 87:1219–1223.

117. Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am J Clin Nutr. 2007; 86:1286–1292.

118. Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysa-ccharides. J Lipid Res. 2009; 50:90–97.

119. Wei X, Yang Z, Rey FE, et al. Fatty acid synthase modulates intestinal barrier function through palmitoylation of mucin 2. Cell Host Microbe. 2012; 11:140–152.

120. Koh YS. Nucleic acid recognition and signaling by toll-like receptor 9: compartment-dependent regulation. J Bacteriol Virol. 2011; 41:131–132.

122. Saberi M, Woods NB, de Luca C, et al. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009; 10:419–429.

123. Kim JM. Inflammatory bowel diseases and inflammasome. Korean J Gastroenterol. 2011; 58:300–310.

124. Vandanmagsar B, Youm YH, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011; 17:179–188.

125. Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011; 145:745–757.

126. Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012; 482:179–185.

127. Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica se-rovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007; 5:2177–2189.

128. Jernberg C, Löfmark S, Edlund C, Jansson JK. Longterm impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010; 156(Pt 11):3216–3223.

129. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009; 15:1183–1189.

130. Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009; 15:653–660.

131. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012; 9:599–608.

132. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008; 105:16731–16736.
133. Jia W, Whitehead RN, Griffiths L, et al. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn's disease? FEMS Microbiol Lett. 2010; 310:138–144.
134. Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012; 9:219–230.

135. Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012; 35:828–838.

136. Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011; 141:1792–1801.

138. Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Philos Soc. 2012; 87:701–730.

139. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012; 6:320–329.

140. Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2011; 9:88–96.

141. Guo B, Harstall C, Louie T, Veldhuyzen van Zanten S, Dieleman LA. Systematic review: faecal transplantation for the treatment of Clostridium difficile-associated disease. Aliment Pharmacol Ther. 2012; 35:865–875.
142. Mattila E, Uusitalo-Seppälä R, Wuorela M, et al. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology. 2012; 142:490–496.
143. Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacter-iotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010; 44:354–360.
Go to : 
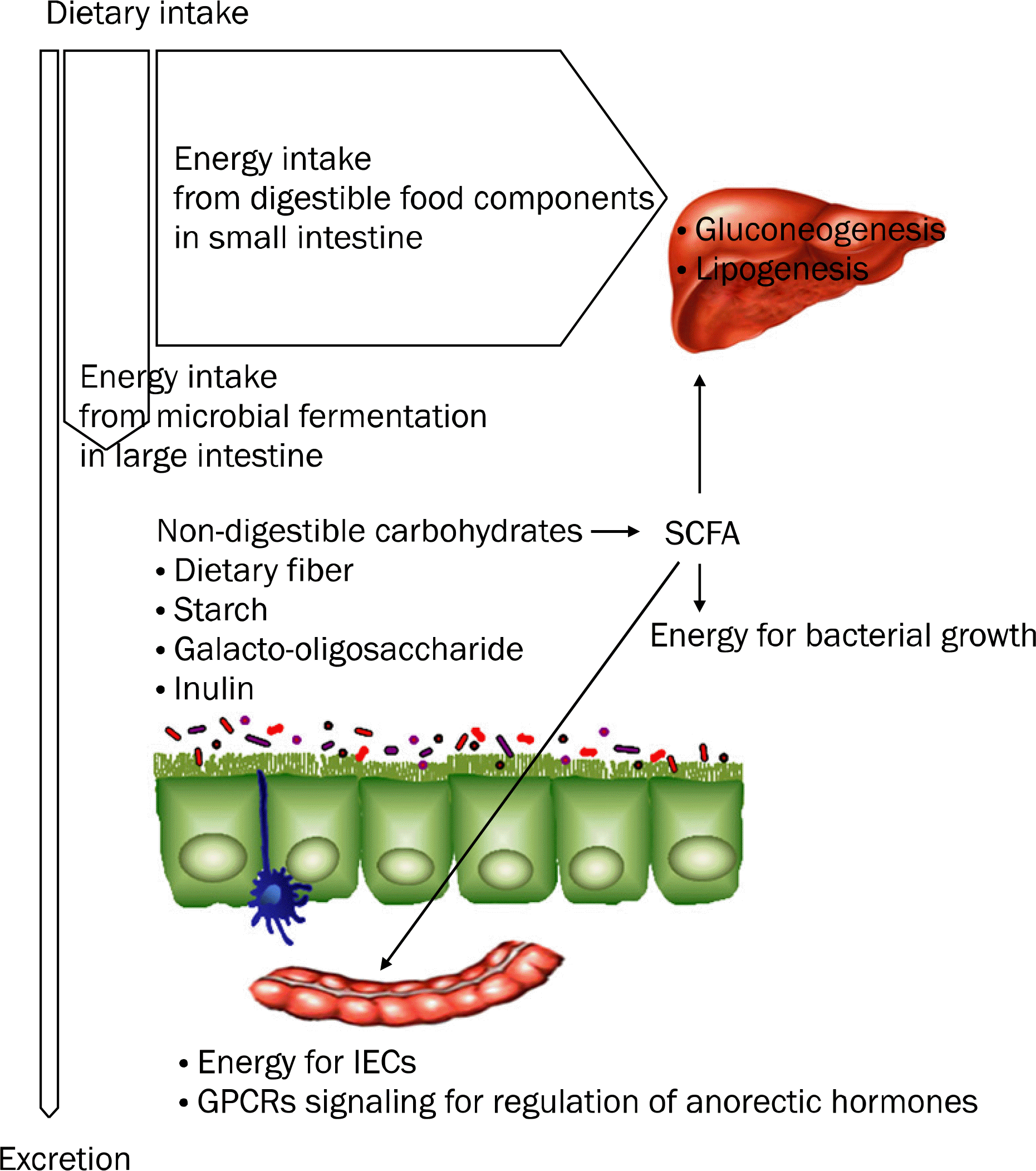 | Fig. 1.Energy intake and fermentation of non-digestible carbohydrates by intestinal microbiota in large intestine. Each gram of glucose that is directly absorbed from the small intestine contributes approximately 3.9 kcal to energy intake. Non-digestible carbohydrates that are resistant to digestion in the small intestine contribute energy indirectly as a result of microbial fermentation in the colon to produce short-chain fatty acids (SCFA) and gases. This contribution to the body's energy intake is approximately 1.5 kcal/g glucose because of the lower energy content of SCFA and their incomplete absorption from the colon. Fermentability depends primarily on the structure of the substrate. However, it may be influenced by methods of food preparation and storage, by host physiology, by gut transit, and potentially by the density and species composition of the intestinal microbiota. Fermentation of non-dige-stible carbohydrates by anaerobic bacteria in the large intestine enables the recovery of only a fraction of the initial energy content for microbial growth. SCFA such as butyrate, acetate, and propionate are absorbed in the colon and butyrate provides energy for intestinal epithelial cells (IECs). Acetate and propionate reach the liver and peripheral organs where they are substrates for gluconeogenesis and lipogenesis. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download