Abstract
Background/Aims
Intraductal papillary mucinous neoplasms (IPMN) are precursor lesions of fatal pancreatic cancer. Physiological function of microRNA is to regulate the stability and translation of mRNA. The aberrant microRNA expression is commonly observed in many cancers. The aim of this study was to analyze the expression pattern of microRNA in IPMN and evaluate the role of the microRNA.
Methods
Using two paraffin-embedded IPMN tissues, microRNA expression of normal tissue, IPMN adenoma and carcinoma were compared by cDNA-mediated annealing, selection, extension and ligation microarray assay. Using real time PCR, expression levels of aberrantly up-regulated microRNAs were assessed in another 20 IPMNs, four pancreatic cancer cell lines (Panc1, MiaPaCa-2, XPA-3, BxPC-3) and immortalized pancreatic ductal cell line (HPNE). Effect of suppressing highly over-expressed two microRNAs in pancreatic cancer cell lines with anti-microRNA inhibitors were evaluated using CCK-8 assay.
Results
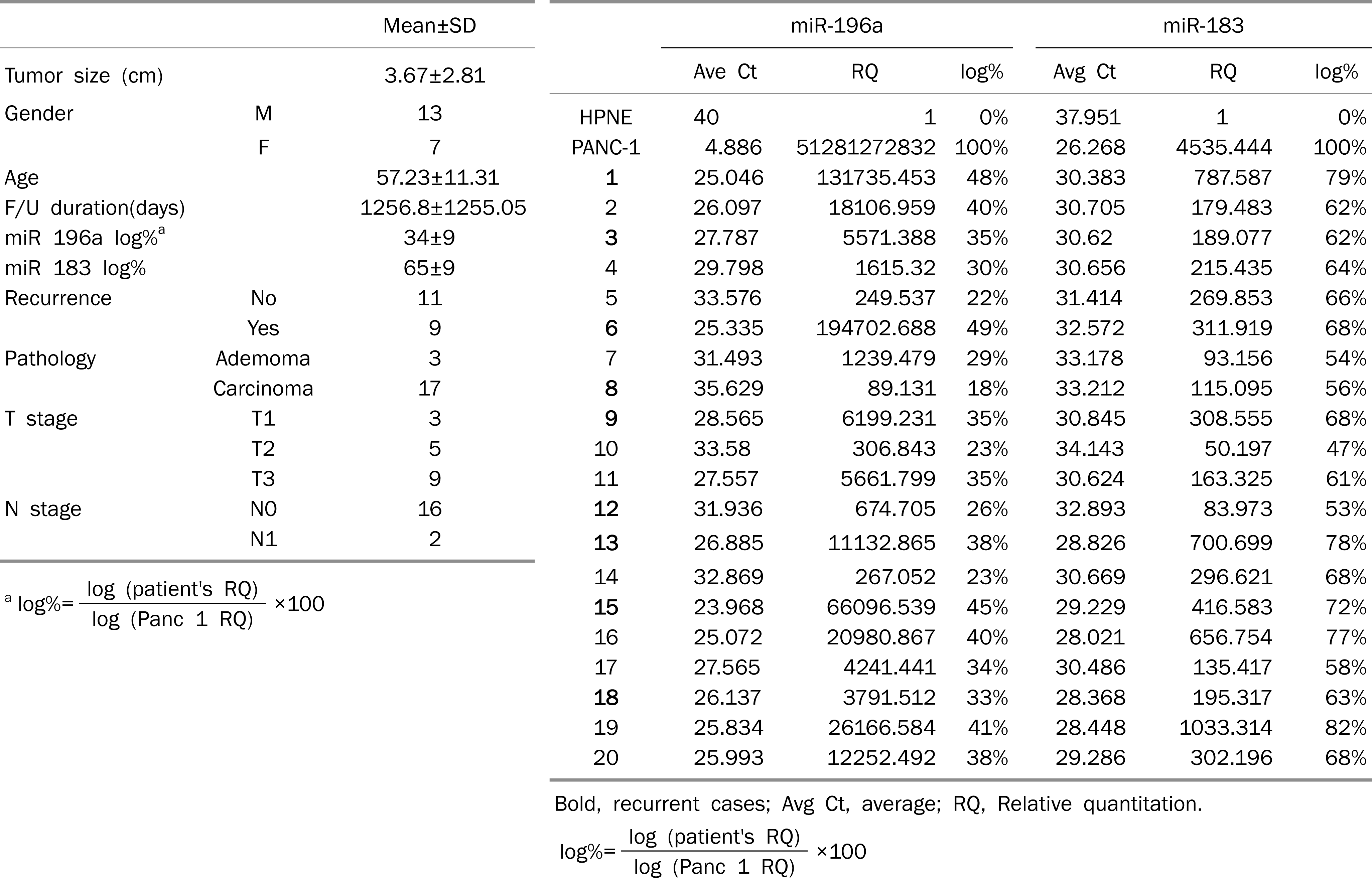
Among aberrantly expressed 122 microRNAs in IPMN, miR-552, miR-25*, miR-183, miR-1300, miR-196a, miR-182*, and miR-30c–1* were consistently increased more than 3-fold. On average, miR-196a and miR-183 increased 10,824 folds and 26,519 folds in four pancreatic cancer cell lines compared with HPNE. These two microRNAs were also over-expressed in 20 IPMNs compared with HPNE. After applying anti-miRNA inhibitors, cell survival of four pancreatic cancer cell lines decreased by 24.5% with anti-miR-196a and by 14.2% with anti-miR-183 on average.
Conclusions
Aberrant expression of 122 microRNAs was observed in IPMN. Two microRNAs, miR-196a and miR-183-increased in IPMN and pancreatic cancer cell lines compared with immortalized dancreatic ductal cell line. The inhibitions of these microRNAs repressed cell proliferation of pancreatic cancer cell lines.
Go to : 
References
3. Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006; 20:211–226.

4. Rosty C, Goggins M. Early detection of pancreatic carcinoma. Hematol Oncol Clin North Am. 2002; 16:37–52.

5. Hruban RH, Wilentz RE, Maitra A. Identification and analysis of precursors to invasive pancreatic cancer. Methods Mol Med. 2005; 103:1–13.

6. Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005; 12:81–91.

7. Kim MH, Lee SK, Chung YH, et al. A case of mucinous ductal ectasia of the pancreas. Korean J Gastroenterol. 1992; 24:160–164.
8. Nagai E, Ueki T, Chijiiwa K, Tanaka M, Tsuneyoshi M. Intraductal papillary mucinous neoplasms of the pancreas associated with so-called “mucinous ductal ectasia”. Histochemical and immunohistochemical analysis of 29 cases. Am J Surg Pathol. 1995; 19:576–589.

9. Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005; 438:685–689.

10. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838.

11. Scherr M, Venturini L, Battmer K, et al. Lentivirus-mediated an-tagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007; 35:e149.

12. He L, He X, Lowe SW, Hannon GJ. MicroRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007; 7:819–822.
14. Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006; 6:259–269.

15. Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formal-in-fixed, paraffin-embedded tissues. J Mol Diagn. 2008; 10:203–211.

16. Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008; 10:513–519.

17. Mittempergher L, de Ronde JJ, Nieuwland M, et al. ene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One. 2011; 6:e17163.
18. Reinholz MM, Eckel-Passow JE, Anderson SK, et al. Expression profiling of formalin-fixed paraffin-embedded primary breast tumors using cancer-specific and whole genome gene panels on the DASLⓇ platform. BMC Med Genomics. 2010; 3:60.

19. Kibriya MG, Jasmine F, Roy S, Paul-Brutus RM, Argos M, Ahsan H. Analyses and interpretation of whole-genome gene expression from formalin-fixed paraffin-embedded tissue: an illustration with breast cancer tissues. BMC Genomics. 2010; 11:622.

20. Waddell N, Cocciardi S, Johnson J, et al. Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-DASL assay. J Pathol. 2010; 221:452–461.

21. Bibikova M, Yeakley JM, Wang-Rodriguez J, Fan JB. Quantitative expression profiling of RNA from formalin-fixed, paraf-fin-embedded tissues using randomly assembled bead arrays. Methods Mol Biol. 2008; 439:159–177.

22. Abramovitz M, Ordanic-Kodani M, Wang Y, et al. Optimization of RNA extraction from FFPE tissues for expression profiling in the DASL assay. Biotechniques. 2008; 44:417–423.

23. Ravo M, Mutarelli M, Ferraro L, et al. Quantitative expression profiling of highly degraded RNA from formalin-fixed, paraf-fin-embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest. 2008; 88:430–440.

24. Bibikova M, Talantov D, Chudin E, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004; 165:1799–1807.

25. Fan JB, Yeakley JM, Bibikova M, et al. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004; 14:878–885.

26. Qiu R, Liu Y, Wu JY, Liu K, Mo W, He R. Misexpression of miR-196a induces eye anomaly in Xenopus laevis. Brain Res Bull. 2009; 79:26–31.

27. Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010; 10:502.

28. Habbe N, Koorstra JB, Mendell JT, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009; 8:340–346.

29. Chien J, Fan JB, Bell DA, et al. Analysis of gene expression in stage I serous tumors identifies critical pathways altered in ovarian cancer. Gynecol Oncol. 2009; 114:3–11.

30. Hammoud ZT, Badve S, Zhao Q, et al. Differential gene expression profiling of esophageal adenocarcinoma. J Thorac Cardiovasc Surg. 2009; 137:829–834.

31. Haller AC, Kanakapalli D, Walter R, Alhasan S, Eliason JF, Everson RB. Transcriptional profiling of degraded RNA in cryopreserved and fixed tissue samples obtained at autopsy. BMC Clin Pathol. 2006; 6:9.

32. Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008; 27:6667–6678.

33. Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia- dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009; 174:1940–1948.
Go to : 
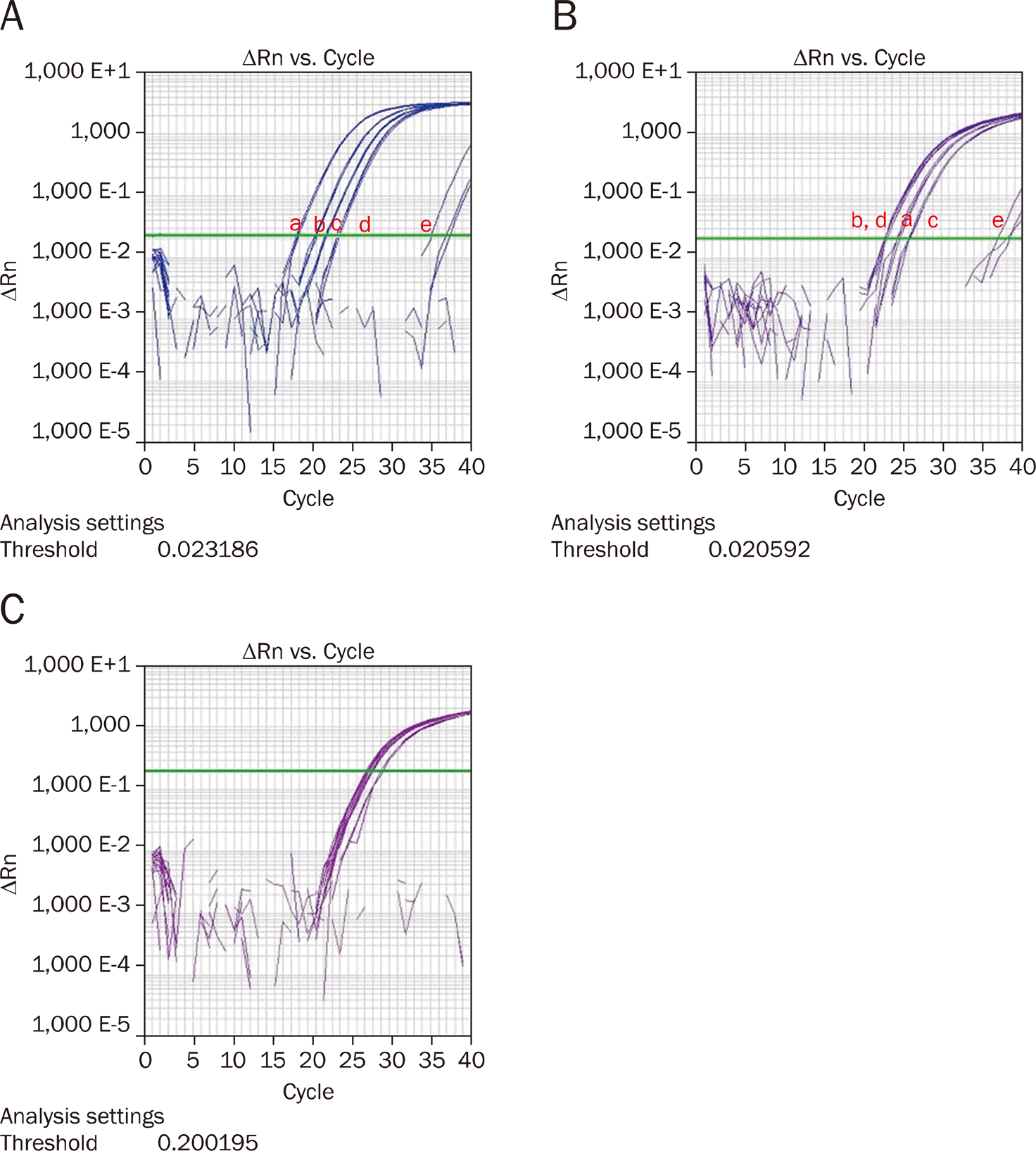 | Fig. 1.MicroRNA-196a (A), microRNA-183 (B), and RNU6 (C) expression in pancreatic cancer cell lines and immortalized pancreatic ductal cells measured by real-time polymerase reaction (a: Panc-1, b: XPA-3, c: BxPC-3, d: MiaPaca-2, e: HPNE). |
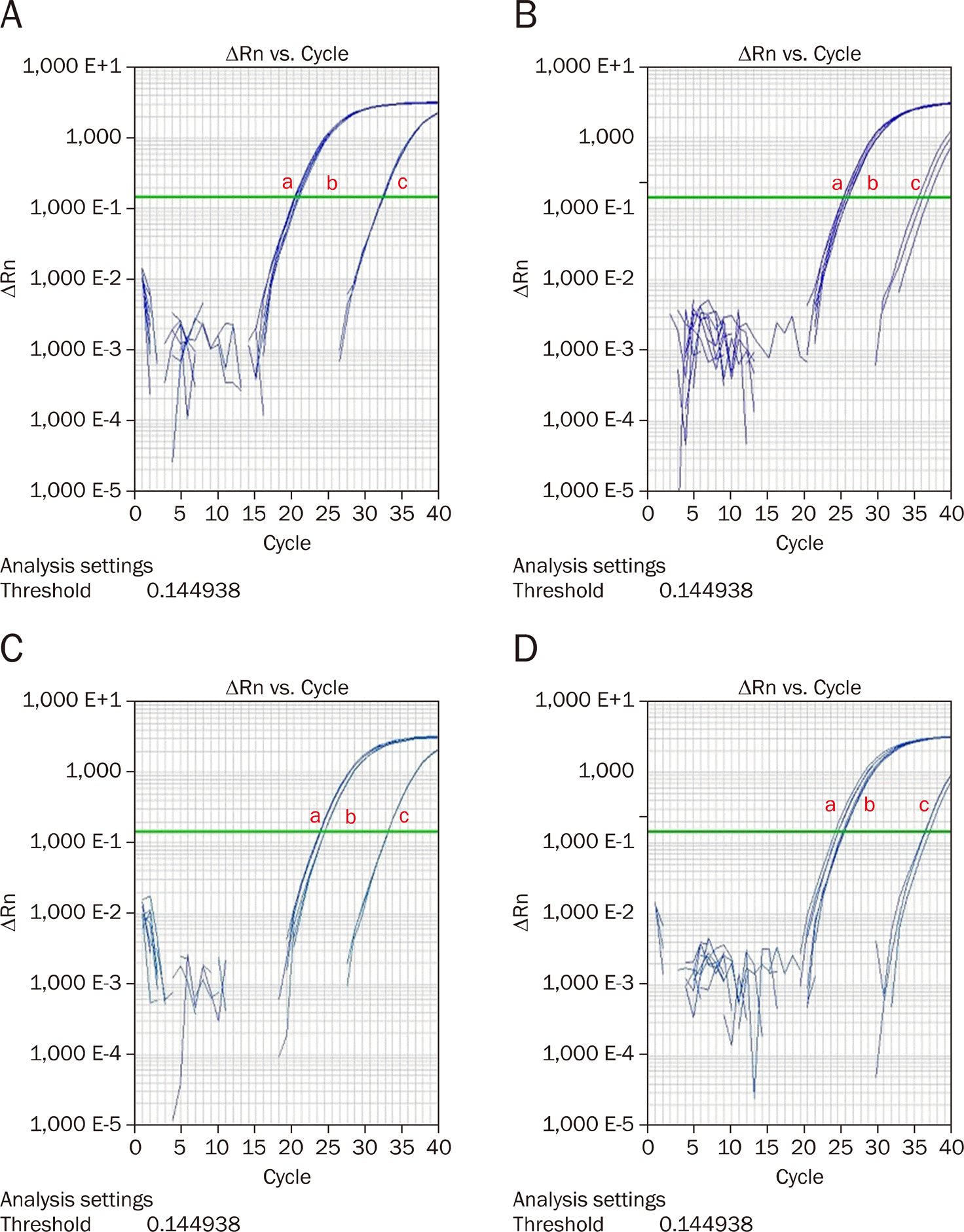 | Fig. 2.MicroRNA-196a suppression in pancreatic cancer cell lines with RNU6B (a: Control, b: With mock, c: With anti-miR-196). (A) Panc-1, (B) MiaPaCa-2, (C) XPA-3, (D) BxPC-3. |
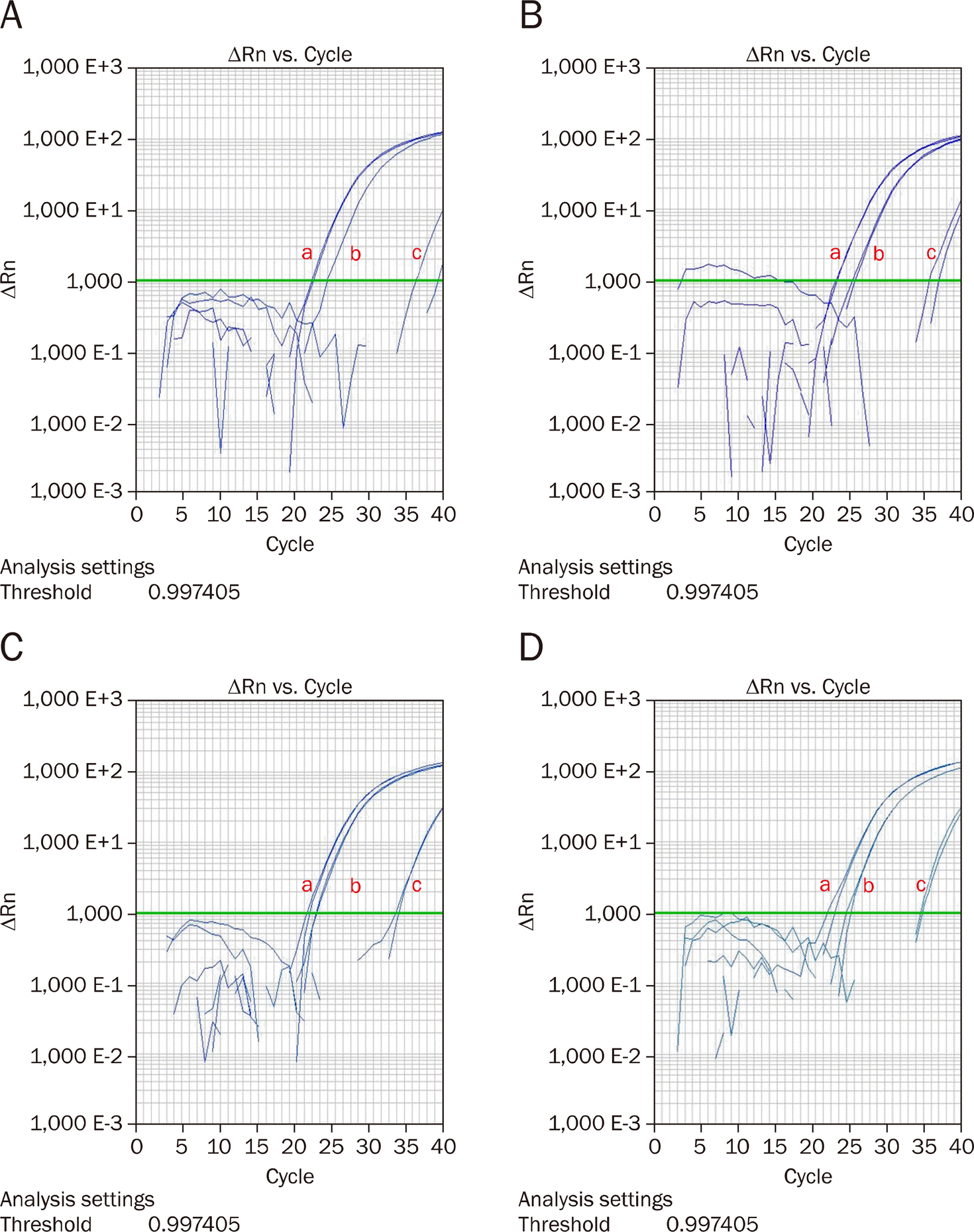 | Fig. 3.MicroRNA-183 suppression in pancreatic cancer cell lines with RNU6B (a: Control, b: With mock, c: With anti-miR-183). (A) Panc-1, (B) MiaPaCa-2, (C) XPA-3, (D) BxPC-3. |
 | Fig. 4.Effect of anti-microRNA-196a and anti-microRNA-183 transfection into Gemcitabine treated pancreatic cancer cell lines. (A) Panc-1, (B) MiaPaCa-2, (C) XPA-3, (D) BxPC-3. |
Table 1.
Up and Down Regulated MicroRNA in IPMN Detected by DASL assay
Table 2.
MicroRNA Expression Patterns in Pancreatic Cancer Cell Lines (Panc-1, MiaPaCa-2, XPA-3, BxPC-3) Comparing with Immortalized Pancreatic Ductal Cell Lline (HPNE) by Real-time Polymerase Chain Reaction
Table 3.
MicroRNA 196a and MicroRNA-183 Expressions in 20 IPMN Patients (A) Patient's Characteristics (B) MicroRNA-196a and Mic




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download