Abstract
Background/Aims
Tetraploid cells are frequently observed in the inflamed mucosal epithelial cells of the patients with Barrett's esophagus or chronic ulcerative colitis. Polyploidy often occurs during cell fusion, abortive cell cycle, and endoreplication. Most tetraploid cells are engaged to apoptotic pathway, but some remaining stable tetraploid cells consequently cause aneuploid-ization and chromosomal instability. We investigated whether tetraploid cells could acquire survival advantage and hold a dominant position for natural selection.
Methods
We established tetraploid cell line (HCT116GH) from parental diploid colorectal cancer cell line (HCT116) via PEG-medi-ated cell fusion and compared its cell viability, cell cycle response and apoptotic fractions responded to H2 O2 with diploid HCT116 and p53 suppressed HCT116/H6 cell lines.
Results
Using MTT assay, plating efficiency and clonogenicity, we evaluated the survival of each cell line. Tetraploid cell line HCT116GH demonstrated an 83 fold greater resistance to 100 μ M H2 O2 than the parental diploid HCT116, and 6 fold greater than even the p53 negative diploid HCT116/E6. Cellular sensitivity, G2/M arrests, and apoptotic proportion were observed less in response to H2 O2 in HCT116GH compared with HCT116 and HCT116/E6. HCT116GH expressed lower level of p53 and p21 than diploid HCT116.
Conclusions
Stable tetraploid cell lines showed enhanced viability in comparison to parental diploid cell lines. The enhanced viability observed in tetraploidization surpassed that from downregulation of p53. Frequent appearance of tetraploid cells in stressful condition can be caused by natural selection owing to their enhanced viability and may consequently contribute to cancer cell transformation.
Go to : 
References
1. Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004; 5:45–54.

2. Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005; 437:1043–1047.

3. Chaves P, Crespo M, Ribeiro C, et al. Chromosomal analysis of Barrett's cells: demonstration of instability and detection of the metaplastic lineage involved. Mod Pathol. 2007; 20:788–796.

4. Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic loss-es, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc Natl Acad Sci U S A. 1996; 93:7081–7084.

5. Sudbø J, Lippman SM, Lee JJ, et al. The influence of resection and aneuploidy on mortality in oral leukoplakia. N Engl J Med. 2004; 350:1405–1413.

6. Levine DS, Rabinovitch PS, Haggitt RC, et al. Distribution of aneuploid cell populations in ulcerative colitis with dysplasia or cancer. Gastroenterology. 1991; 101:1198–1210.

7. Ornitz DM, Hammer RE, Messing A, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987; 238:188–193.

8. Olaharski AJ, Sotelo R, Solorza-Luna G, et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006; 27:337–343.

9. Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997; 386:623–627.

10. Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007; 17:157–162.

11. Chang DK, Goel A, Ricciardiello L, et al. Effect of H(2)O(2) on cell cycle and survival in DNA mismatch repair-deficient and -proficient cell lines. Cancer Lett. 2003; 195:243–251.

12. Honma M, Momose M, Tanabe H, et al. Requirement of wild-type p53 protein for maintenance of chromosomal integrity. Mol Carcinog. 2000; 28:203–214.

13. Lechner MS, Laimins LA. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. 1994; 68:4262–4273.

14. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983; 65:55–63.

15. Scudiero DA, Shoemaker RH, Paull KD, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988; 48:4827–4833.
16. Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phos-phatidylserine expression on B cells undergoing apoptosis. Blood. 1994; 84:1415–1420.

17. Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phospha-tidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995; 184:39–51.

18. Parker SB, Eichele G, Zhang P, et al. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995; 267:1024–1027.
20. Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005; 6:567–575.

21. Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002; 21:483–492.

22. Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005; 8:7–12.

Go to : 
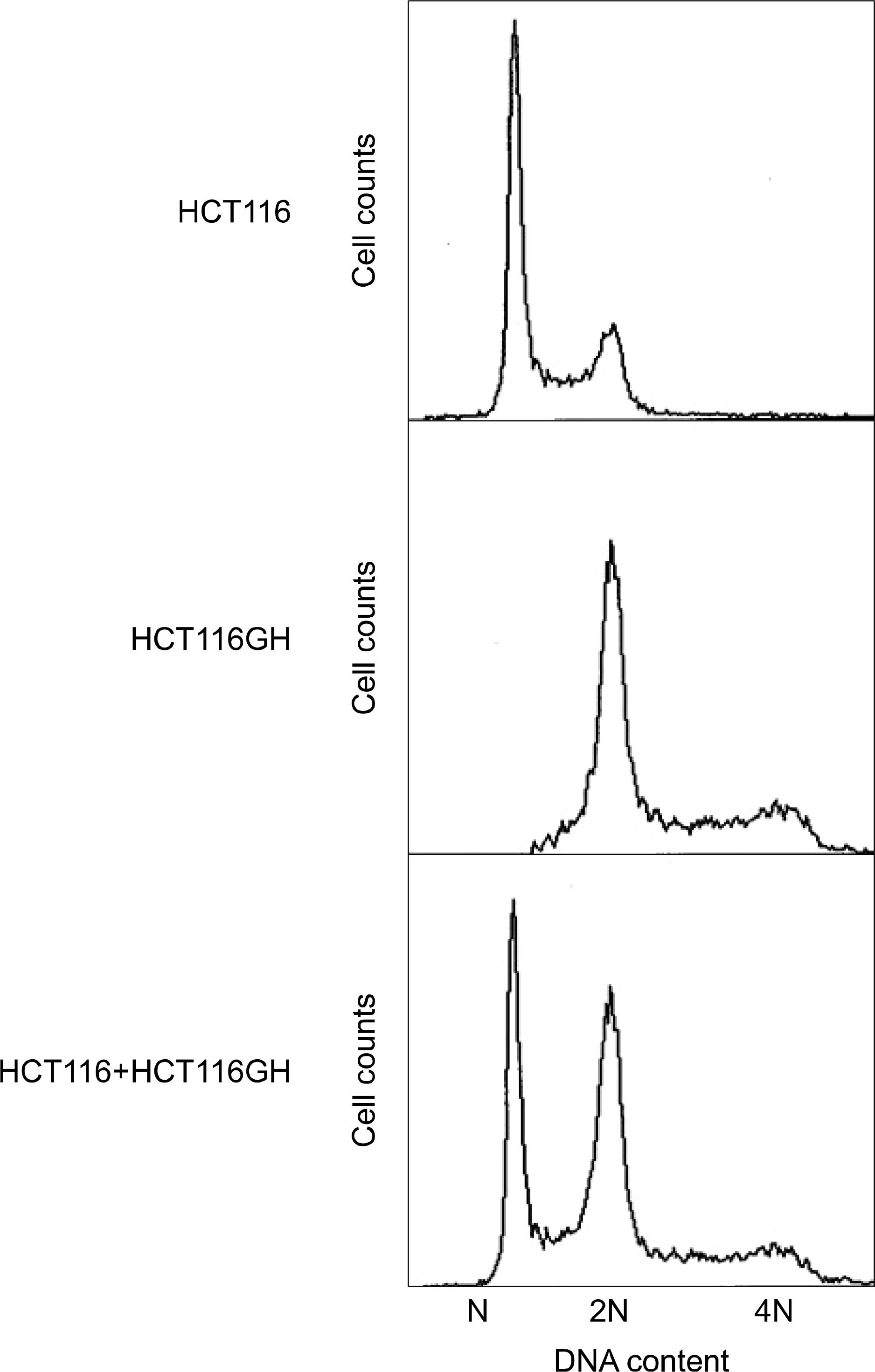 | Fig. 1.Establishment of the tetraploid cell line. Tetraploid cell line HCT116GH demonstrated 2N at G1 and 4N at G2 phase when the parental diploid cell line HCT116 demonstated N at G1 and 2N at G2 phase. |
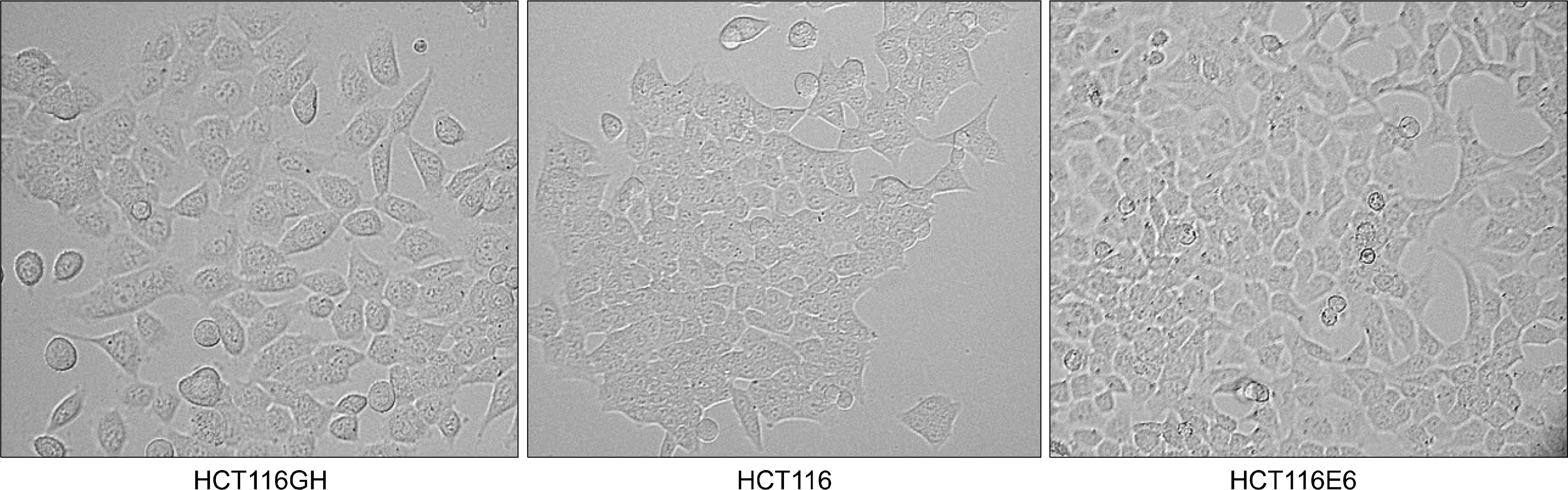 | Fig. 2.Morphology of the tetraploid cells. Tetraploid cell line HCT116GH demonstrated that the size of cells was larger than that of parental diploid HCT116 cells. |
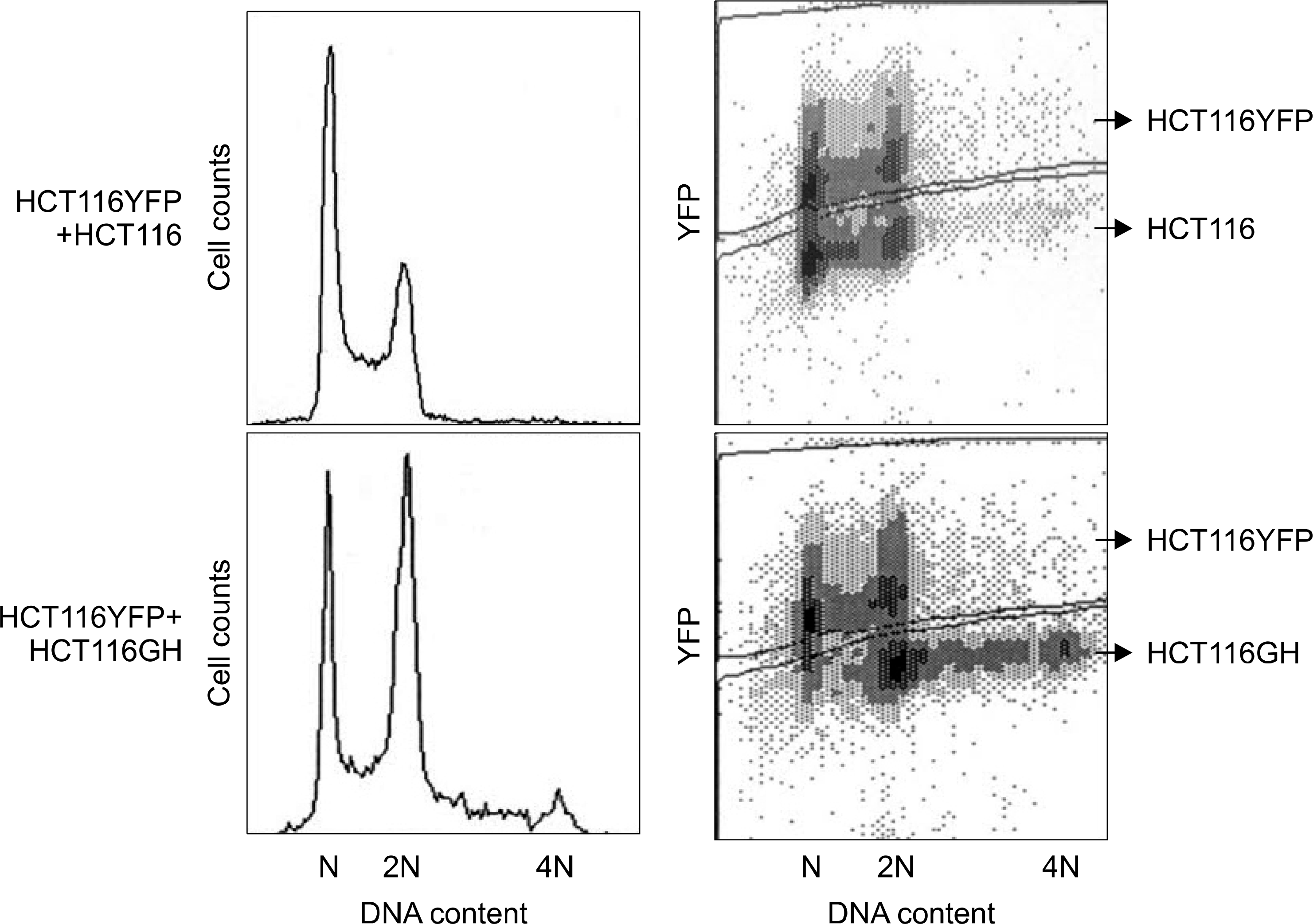 | Fig. 3.Confirmation of stable tetraploid cell line with the reference of the diploid HCT116YFP cell line. In the lower panel depicting the mixture of HCY116YFP and HCT116GH, 2N peak has increased and 4N peak newly appeared, which confirmed that HCT116GH was a stably established tetraploid cell line. |
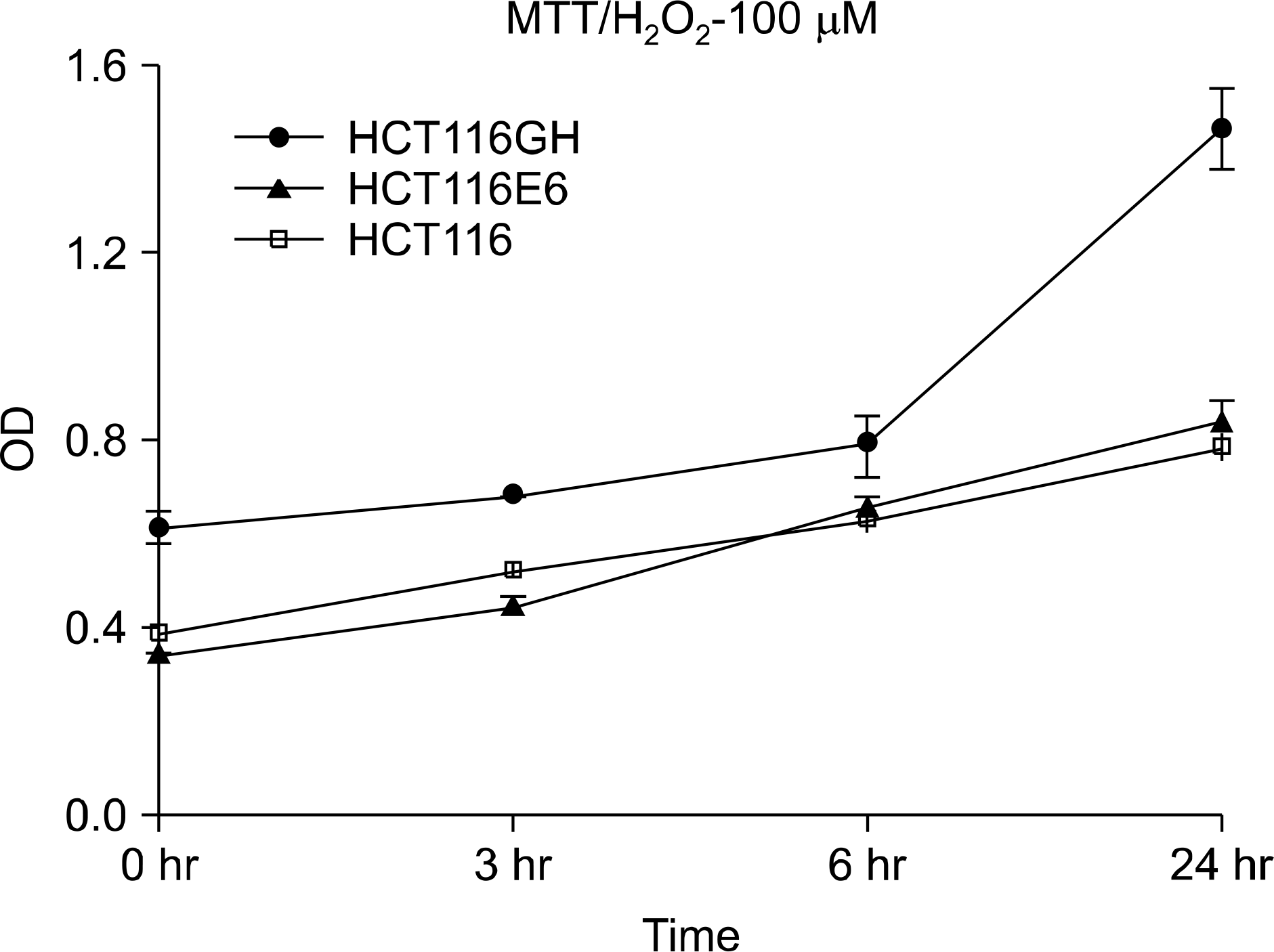 | Fig. 4.Comparison of survival among tetraploid cell line, patental diploid cell line and p53(−) diploid cell line. MTT assay demonstrated that the tetraploid cell line HCT116GH showed higher survival at each time scale after H2O2 challenging (p<0.05, ANOVA). |
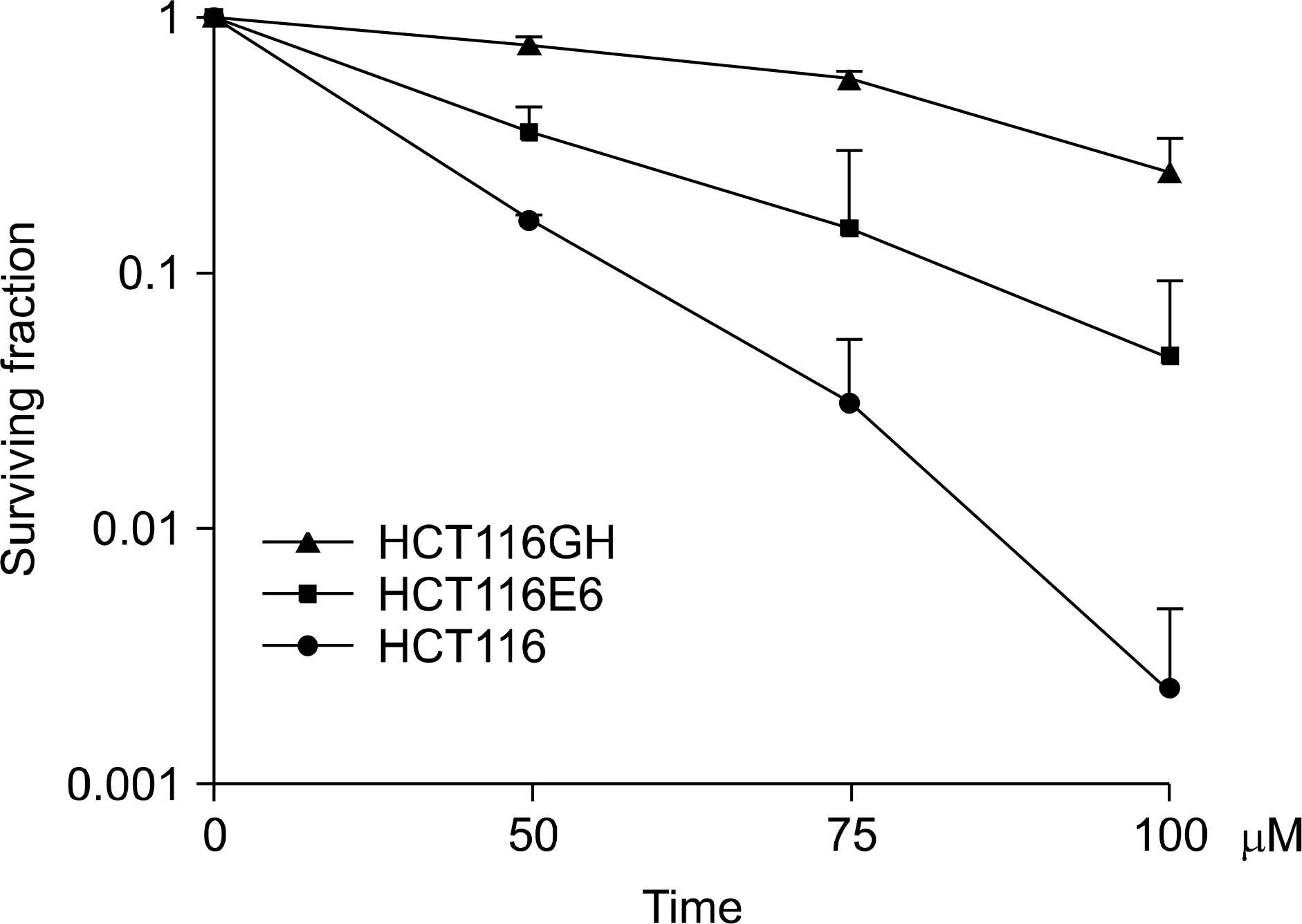 | Fig. 5.Comparison of the plating efficiency between tetraploid cell line, patental diploid cell line and p53(−) diploid cell line. Plating efficiency demonstrated that tetraploid cell line HCT116GH was resistant to H2 O2 in comparison to the parental diploid HCT116 and even p53 negative diploid cell line HCT116/E6 (p<0.05, ANOVA). |
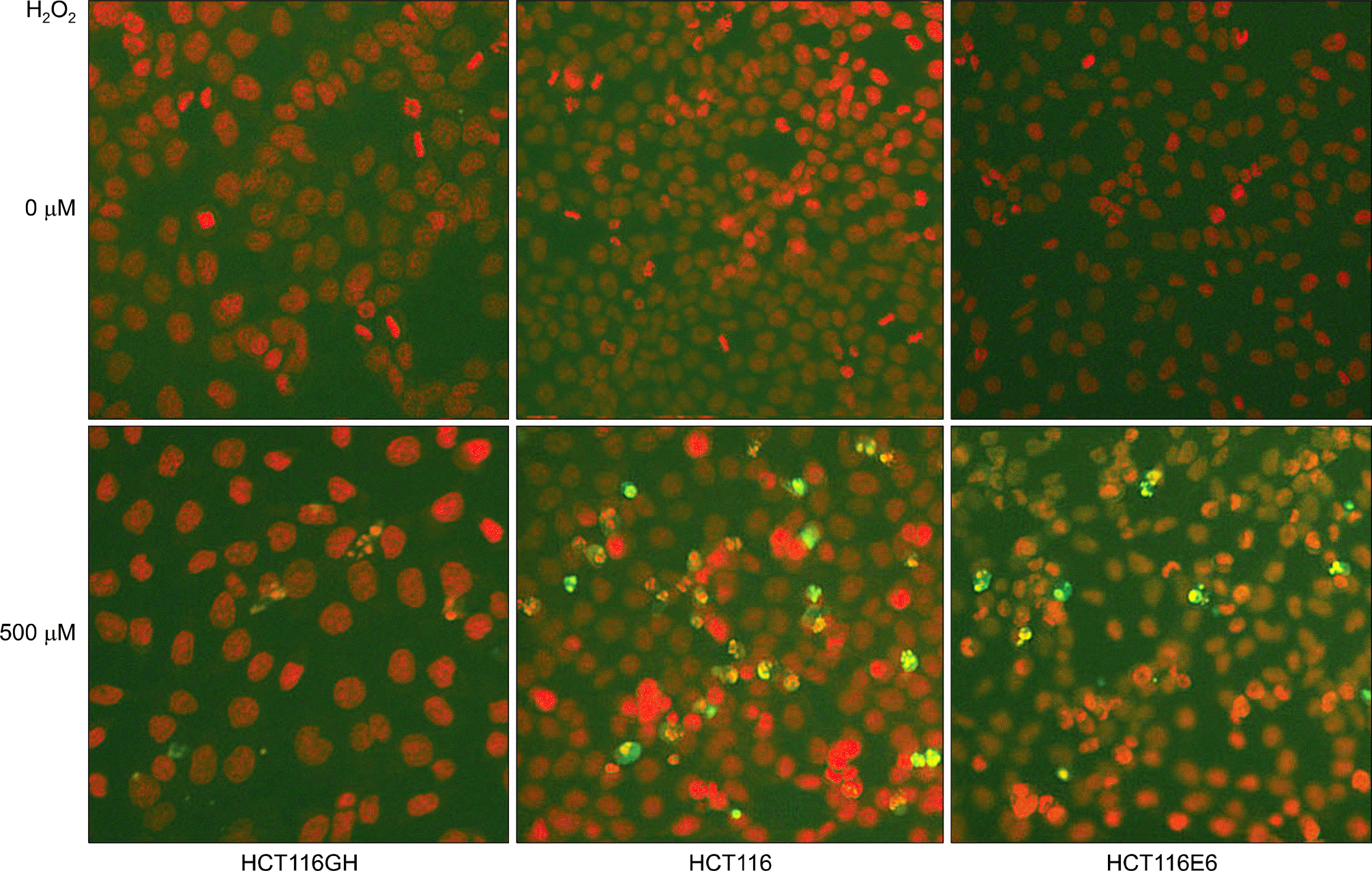 | Fig. 6.Comparison of the apoptosis to oxyradical stress among tetraploid cell line, parental diploid cell line and p53(−) diploid cell line. Apoptotic fraction assessed by annexin-V staining was lowest in the tetraploid cell line HCT116GH. |
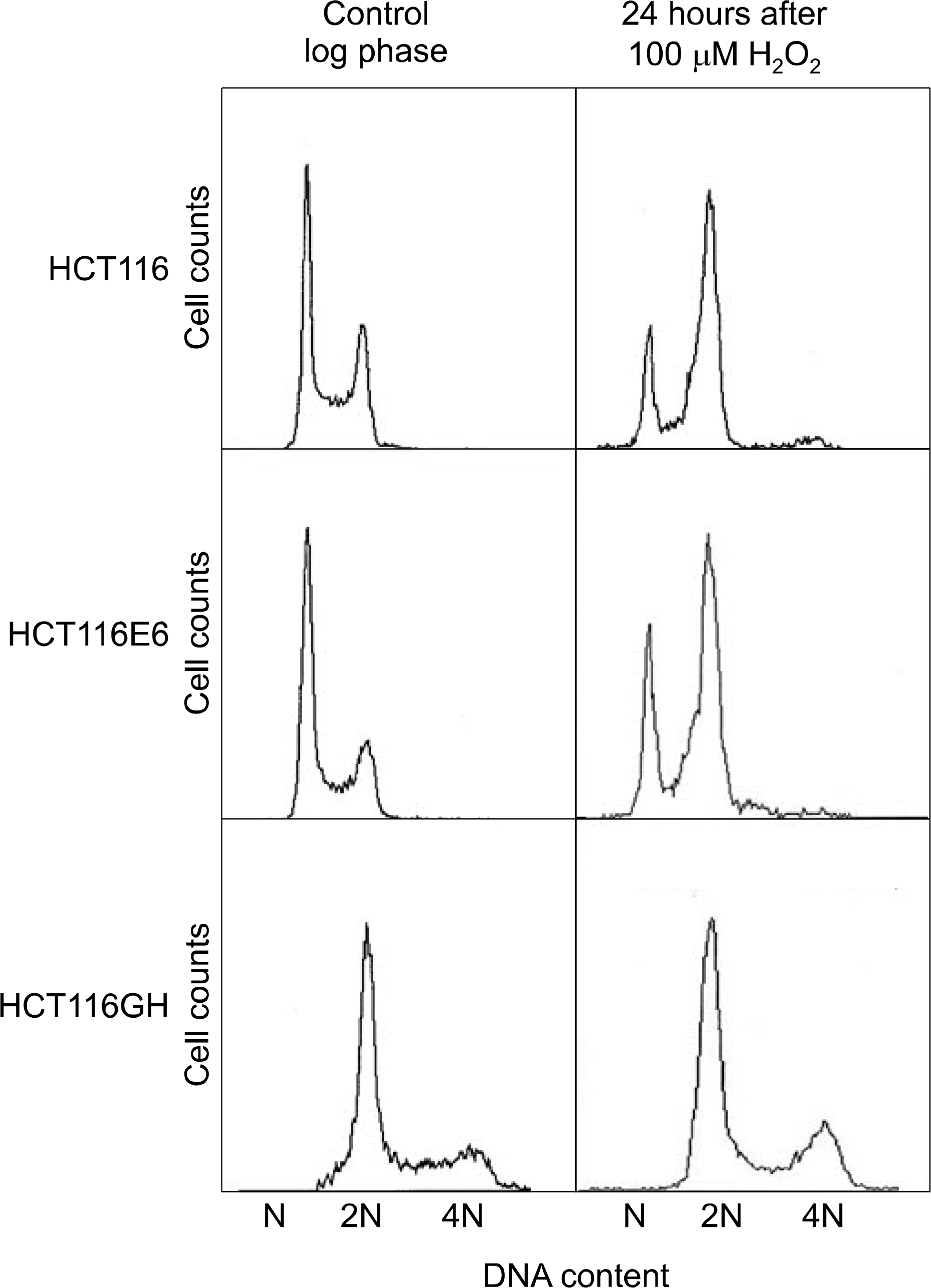 | Fig. 7.Comparison of the G2/M arrest status to oxyradical stress among tetraploid cell line, patental diploid cell line and p53(−) diploid cell line. The fraction of cells in G2/M arrest was lowest in the tetraploid cell line HCT116GH. |
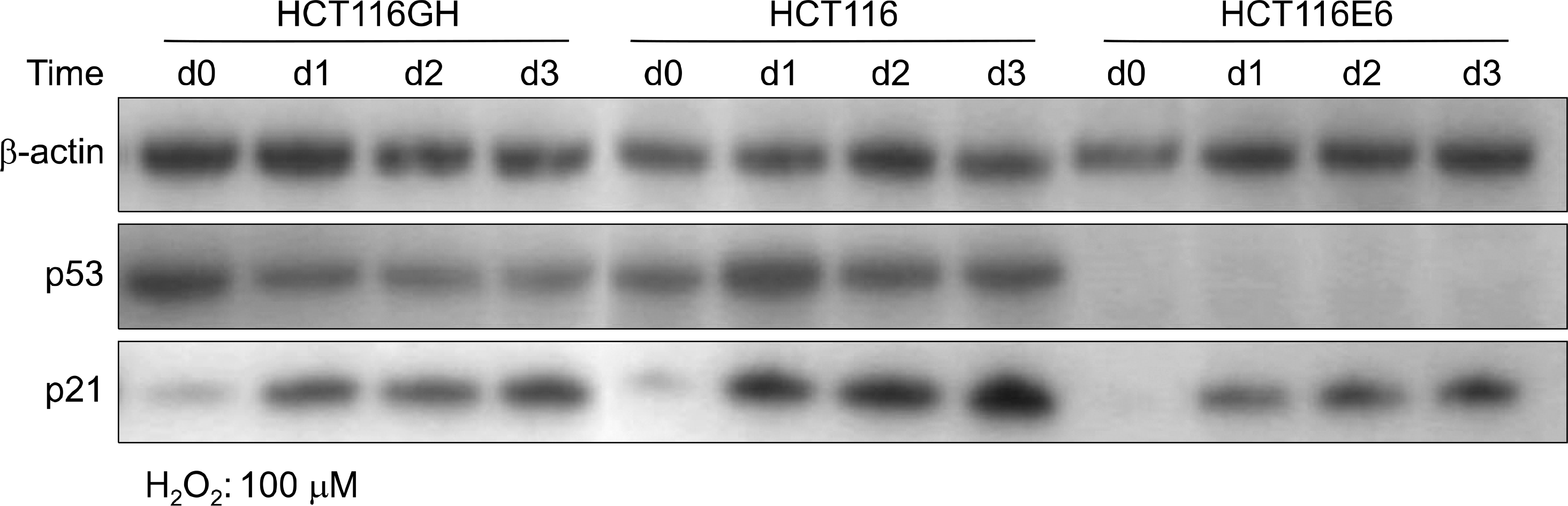 | Fig. 8.Expression of p53 and p21 to oxyradical stress in tetraploid cell line, patental diploid cell line and p53(−) diploid cell line. Tetraploid cell line HCT116GH expressed lower level of p53 and p21 than diploid HCT116. p53 negative HCT116E6 expressed p21 less abundantly than HCT116, with p53 independent manner. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download