Abstract
Background/Aims
Saccharomyces boulardii has been reported to be beneficial in the treatment of inflammatory bowel disease. The aim of this work was to evaluate the effect of S. boulardii in a mice model of 2,4,6-trini-trobencene sulfonic acid (TNBS) induced colitis and analyze the expression of genes in S. boulardii treated mice by microarray.
Methods
BALB/c mice received TNBS or TNBS and S. boulardii treatment for 4 days. Microarray was performed on total mRNA form colon, and histologic evaluation was also performed.
Results
In mice treated with S. boulardii, the histological appearance and mortality rate were significantly restored compared with rats receiving only TNBS. Among 330 genes which were altered by both S. boulardii and TNBS (>2 folds), 193 genes were down-regulated by S. boulardii in microarray. Most of genes which were down-regulated by S. bouardii were functionally classified as inflammatory and immune response related genes.
REFERENCES
1. Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989; 66:365–378.
2. Goossens D, Jonkers D, Stobberingh E, van den Bogaard A, Russel M, Stockbrugger R. Probiotics in gastroenterology: indications and future perspectives. Scand J Gastroenterol. 2003; 239(suppl):S15–S23.
3. Zanello G, Meurens F, Berri M, Salmon H. Saccharomyces boulardii effects on gastrointestinal diseases. Curr Issues Mol Biol. 2009; 11:47–58.
4. Plein K, Hotz J. Therapeutic effects of Saccharomyces boulardii on mild residual symptoms in a stable phase of Crohn's disease with special respect to chronic diarrhea–a pilot study. Z Gastroenterol. 1993; 31:129–134.
5. Guslandi M, Mezzi G, Sorghi M, Testoni PA. Saccharomyces boulardii in maintenance treatment of Crohn's disease. Dig Dis Sci. 2000; 45:1462–1464.
6. Garcia Vilela E, De Lourdes De Abreu Ferrari M, Oswaldo Da Gama Torres H, et al. Influence of Saccharomyces boulardii on the intestinal permeability of patients with Crohn's disease in remission. Scand J Gastroenterol. 2008; 43:842–848.
7. Rodrigues AC, Cara DC, Fretez SH, et al. Saccharomyces boulardii stimulates sIgA production and the phagocytic system of gnotobiotic mice. J Appl Microbiol. 2000; 89:404–414.
8. Caetano JA, Paramés MT, Babo MJ, et al. Immunopharmaco-logical effects of Saccharomyces boulardii in healthy human volunteers. Int J Immunopharmacol. 1986; 8:245–259.
9. Mumy KL, Chen X, Kelly CP, McCormick BA. Saccharomyces boulardii interferes with Shigella pathogenesis by post-invasion signaling events. Am J Physiol Gastrointest Liver Physiol. 2008; 294:599–609.
10. Sougioultzis S, Simeonidis S, Bhaskar KR, et al. Saccharomyces boulardii produces a soluble anti-inflammatory factor that inhibits NF-kappaB-mediated IL-8 gene expression. Biochem Biophys Res Commun. 2006; 343:69–76.
11. Girard P, Pansart Y, Gillardin JM. Inducible nitric oxide synthase involvement in the mechanism of action of Saccharomyces boulardii in castor oil-induced diarrhoea in rats. Nitric Oxide. 2005; 13:163–169.
12. Lee SK, Kim HJ, Chi SG, et al. Saccharomyces boulardii activates expression of peroxisome proliferator-activated receptor-gamma in HT-29 cells. Korean J Gastroenterol. 2005; 45:328–334.
13. Medina C, Videla S, Radomski A, et al. Therapeutic effect of phenantroline in two rat models of inflammatory bowel disease. Scand J Gastroenterol. 2001; 36:1314–1319.
14. Morris GP, Beck PL, Herridge MS, Depew WT, Szewczuk MR, Wallace JL. Hapten-induced model of chronic inflammation and ulceration in the rat colon. Gastroenterology. 1989; 96:795–803.

15. Yamamoto S, Isuzugawa K, Takahashi Y, et al. Intestinal gene expression in TNBS treated mice using genechip and subtractive cDNA analysis: implications for Crohn's disease. Biol Pharm Bull. 2005; 28:2046–2053.

16. Keates AC, Castagliuolo I, Cruickshank WW, et al. Interleukin 16 is up-regulated in Crohn's disease and participates in TNBS colitis in mice. Gastroenterology. 2000; 119:972–982.

17. Spahn TW, Kucharzik T. Modulating the intestinal immune system: the role of lymphotoxin and GALT organs. Gut. 2004; 53:456–465.

18. O'Rourke KP, O'Donoghue G, Adams C, et al. High levels of Lymphotoxin-Beta (LT-Beta) gene expression in rheumatoid arthritis synovium: clinical and cytokine correlations. Rheumatol Int. 2008; 28:979–986.
19. Aggarwal A, Agarwal S, Misra R. Chemokine and chemokine receptor analysis reveals elevated interferon-inducible protein-10 (IP)-10/CXCL10 levels and increased number of CCR5+ and CXCR3+ CD4 T cells in synovial fluid of patients with enthesitis-related arthritis (ERA). Clin Exp Immunol. 2007; 148:515–519.

20. Lee JW, Bajwa PJ, Carson MJ, et al. Fenofibrate represses interleukin-17 and interferon-gamma expression and improves colitis in interleukin-10-deficient mice. Gastroenterology. 2007; 133:108–123.
21. Lacher M, Kappler R, Berkholz S, Baurecht H, von Schwei-nitz D, Koletzko S. Association of a CXCL9 polymorphism with pediatric Crohn's disease. Biochem Biophys Res Commun. 2007; 363:701–707.

22. King NJ, Thomas SR. Molecules in focus: indoleamine 2,3-dioxygenase. Int J Biochem Cell Biol. 2007; 39:2167–2172.

23. Munn DH, Zhou M, Attwood JT, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998; 281:1191–1193.

24. Platten M, Ho PP, Youssef S, et al. Treatment of autoimmune neuroinflammation with a synthetic tryptophan metabolite. Science. 2005; 310:850–855.

25. Brandacher G, Winkler C, Schroecksnadel K, Margreiter R, Fuchs D. Antitumoral activity of interferon-gamma involved in impaired immune function in cancer patients. Curr Drug Metab. 2006; 7:599–612.
Fig. 1.
The effect of S. boulardii on the histological features of TNBS-induced colitis. Representative photographs of colon sections were shown after treatment with vehicle (A), TNBS (B), and TNBS with S. boulardii (C) (H&E staining, ×40). Histological scoring is shown by graph (D). Results are the mean±SD from five rats per group (∗p<0.05). SB, S. boulardii.
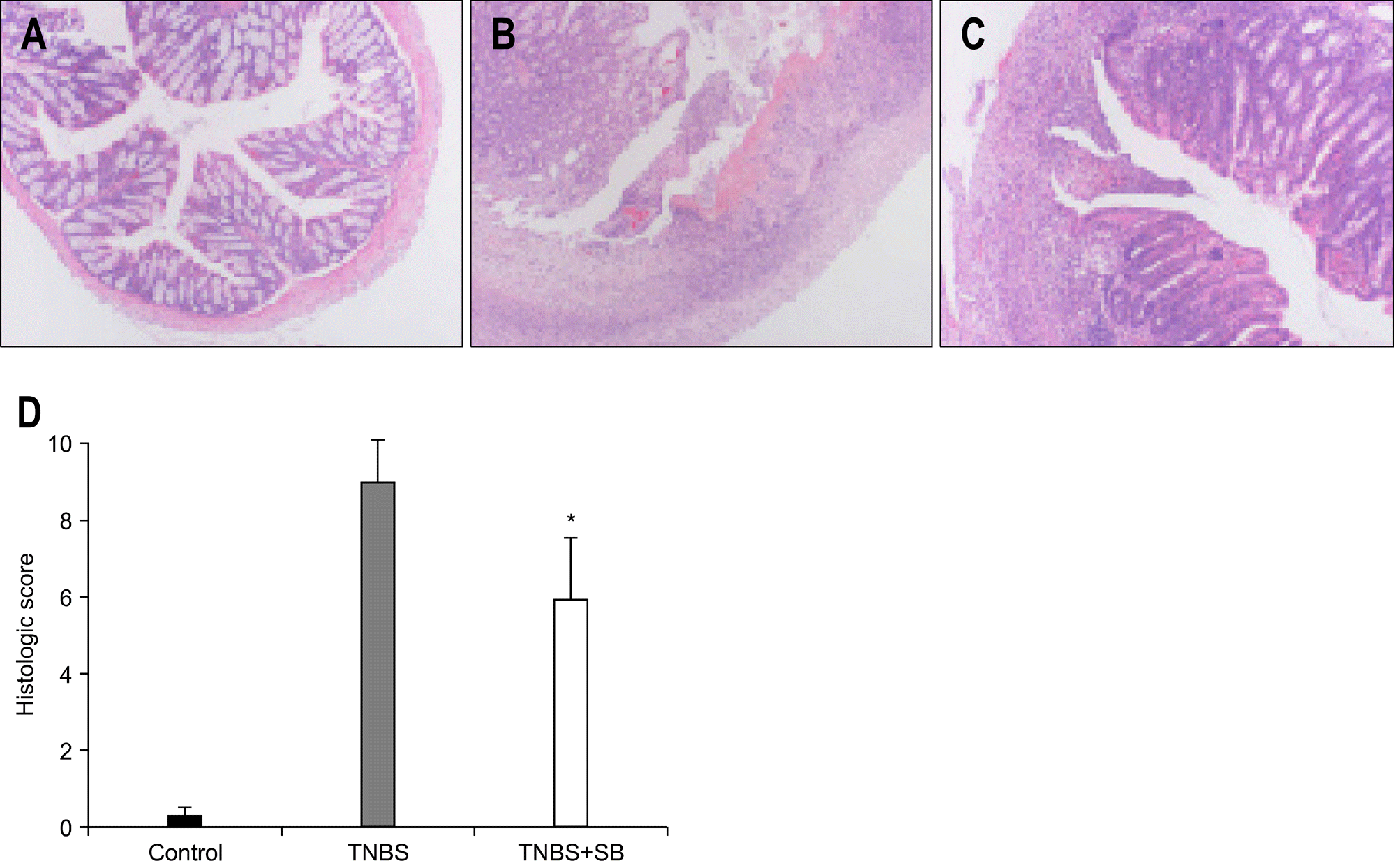
Fig. 2.
Functional classification of differentially expressed genes on treatment of Saccharomyces boulardii.
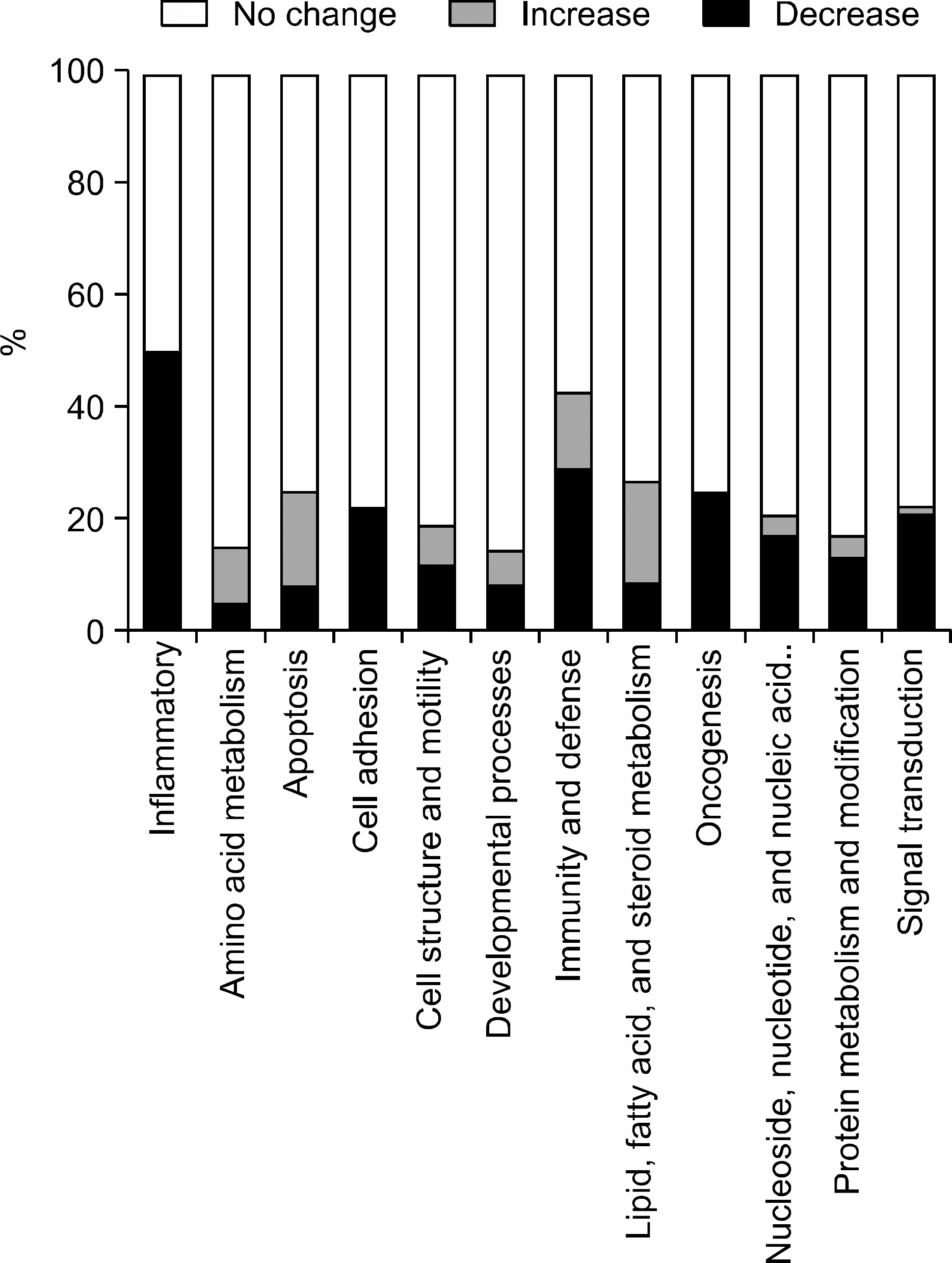
Table 1.
Down-regulated Genes Modulations by S. boulardii on TNBS-induced Colitis Mice
Table 2.
Up-regulated Genes Modulations by S. boulardii on TNBS-induced Colitis Mice




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download