Abstract
Background/Aims
The investigation of a specific tumor marker for hepatocellular carcinoma (HCC) is needed to examine the carcinogenesis and to select the patients for treatment options. The aim of this study was to find the genes related to HCC. We also examined the expression level of these genes in cancer cell lines and tissue specimens.
Methods
Three pairs of HCC tissue and non-neoplastic hepatic tissue around the HCC were collected from three patients who underwent resection for HCC. Differential display reverse transcriptase-PCR (DD RT-PCR) using GeneFishing TM PCR was used to detect the differences in the gene expression between in HCC tissue and non-neoplatic tissue. Up- or down-regulated genes in HCC tissue were identified through BLAST searches after cloning and sequencing assays. Real-time RT-PCR assay was employed to detect the expression rate in 11 HCC tissues and human cancer cell lines.
Go to : 
REFERENCES
1. Park JW, Kim CM. Epidemiology of hepatocellular carcinoma in Korea. Korean J Hepatol. 2005; 11:303–310.
2. Kaino M. Alterations in the tumor suppressor genes p53, RB, p16/MTS1, and p15/MTS2 in human pancreatic cancer and hepatoma cell lines. J Gastroenterol. 1997; 32:40–46.
3. Ashida K, Kishimoto Y, Nakamoto K, et al. Loss of heterozygosity of the retinoblastoma gene in liver cirrhosis accom-panying hepatocellular carcinoma. J Cancer Res Clin Oncol. 1997; 123:489–495.

4. Kondoh N, Wakatsuki T, Ryo A, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999; 59:4990–4996.
5. Song JY, Choi JH, Lee KJ, et al. Identification of the gene associated with hepatocellular carcinoma using differential gene expresion. Korean J Hepatol. 2001; 7:265–272.
6. Cui XS, Shin MR, Lee KA, Kim NH. Identification of differentially expressed genes in murine embryos at the blastocyst stage using annealing control primer system. Mol Reprod Dev. 2005; 70:278–287.

7. Okabe H, Satoh S, Kato T, et al. Genomewide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001; 61:2129–2137.
8. Jia HL, Ye QH, Qin LX, et al. Gene expression profiling re-veals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007; 13:1133–1139.

9. Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007; 27:55–76.

10. Wang ZX, Wang HY, Wu MC. Identification and characterization of a novel human hepatocellular carcinoma-associated gene. Br J Cancer. 2001; 85:1162–1167.

11. Nomura F, Yaguchi M, Togawa A, et al. Enhancement of poly-adenosine diphosphate- ribosylation in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2000; 15:529–535.
12. Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, et al. Apoptosis-inducing factor mediates poly(ADP-ri-bose) (PAR) polymer-induced cell death. PNAS. 2006; 103:18314–18319.

13. Westra R, Jansen TL, Gouw AS, de Jong KP. Anti-nucleair antibody (ANA) positivity caused by paraneoplastic antibodies due to abundant p53 expression in early hepatic carcinoma. Neth J Med. 2003; 61:300–303.
14. Jaskiewicz K. Chasen MR, Robson SC. Differential expression of extracellular matrix proteins and integrins in hepatocellular carcinoma and chronic liver disease. Anticancer Res. 1993; 13:2229–2237.
15. Moore S, Knudsen B, True LD, et al. Loss of stearoyl-CoA desaturase expression is a frequent event in prostate carcinoma. Int J Cancer. 2005; 114:563–571.

16. Falvella FS, Pascale RM, Gariboldi M, et al. Stearoyl-CoA desaturase 1 (SCD1) gene overexpression is associated with genetic predisposition to hepatocarcinogenesis in mice and rats. Carcinogenesis. 2002; 23:1933–1936.

17. Jiang J, Nilsson-Ehle P, Xu N. Influence of liver cancer on lipid and lipoprotein metabolism. Lipids Health Dis. 2006; 5:4.

18. Yahagi N, Shimano H, Hasegawa K, et al. Co-ordinate acti-vation of lipogenic enzymes in hepatocellular carcinoma. Eur J Cancer. 2005; 41:1316–1322.

19. Huang J, Zhang X, Zhang M, et al. Upregulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma. Carcinogenesis. 2007; 28:1094–1103.

20. Lencioni RA, Allgaier HP, Cioni D, et al. Small hepatocellular carcinoma in cirrhosis: randomized comparison of radio-frequency thermal ablation versus percutaneous ethanol injection. Radiology. 2003; 228:235.

21. Liao C, Zhao M, Song H, Uchida K, Yokoyama KK, Li T. Identification of the gene for a novel liver-related putative tumor suppressor at a high-frequency loss of heterozygosity region of chromosome 8p23 in human hepatocellular carcinoma. Hepatology. 2000; 32:721–727.

22. Shirahashi H, Sakaida I, Terai S, Hironaka K, Kusano N, Okita K. Ubiquitin is a possible new predictive marker for the recurrence of human hepatocellular carcinoma. Liver. 2002; 22:413–418.
Go to : 
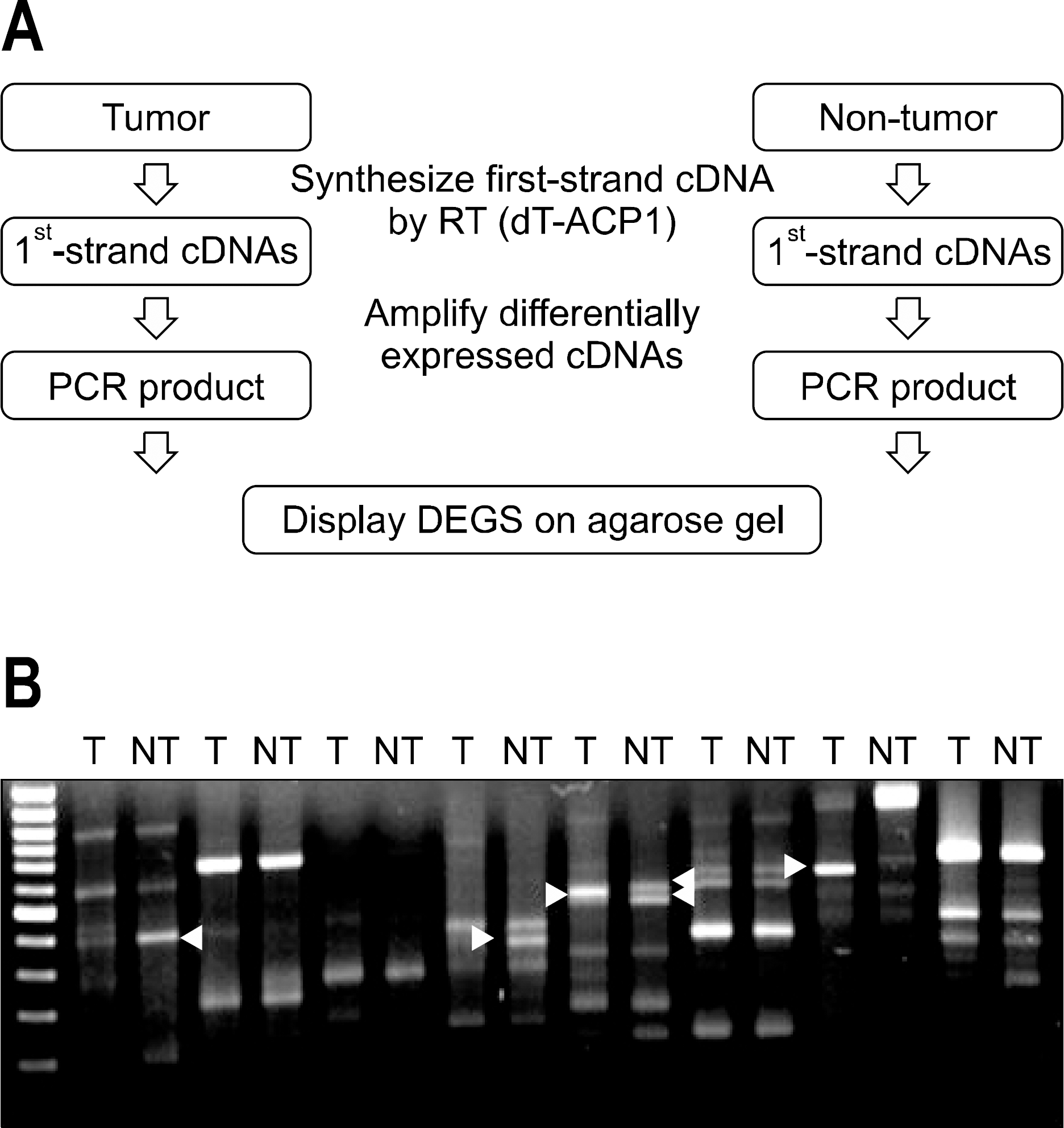 | Fig. 1.Annealing control primer (ACP)-based polymerase chain reaction (PCR) for the identification of differentially expressed genes (DEG) from tumor tissue (T) and non tumor tissues (NT). Total RNA extracted from T and NT were used for the syn-thesis of 1st strand cDNA using dT-ACP1 primer. 2nd strand cDNAs were amplified during second-stage PCR using a combi-nation of dT-ACP2 and one of 120 arbitrary ACP primers (A). Products were separated on an agarose gel to identify the differentially expressed genes that were highly expressed in T and NT tumor liver (B). |
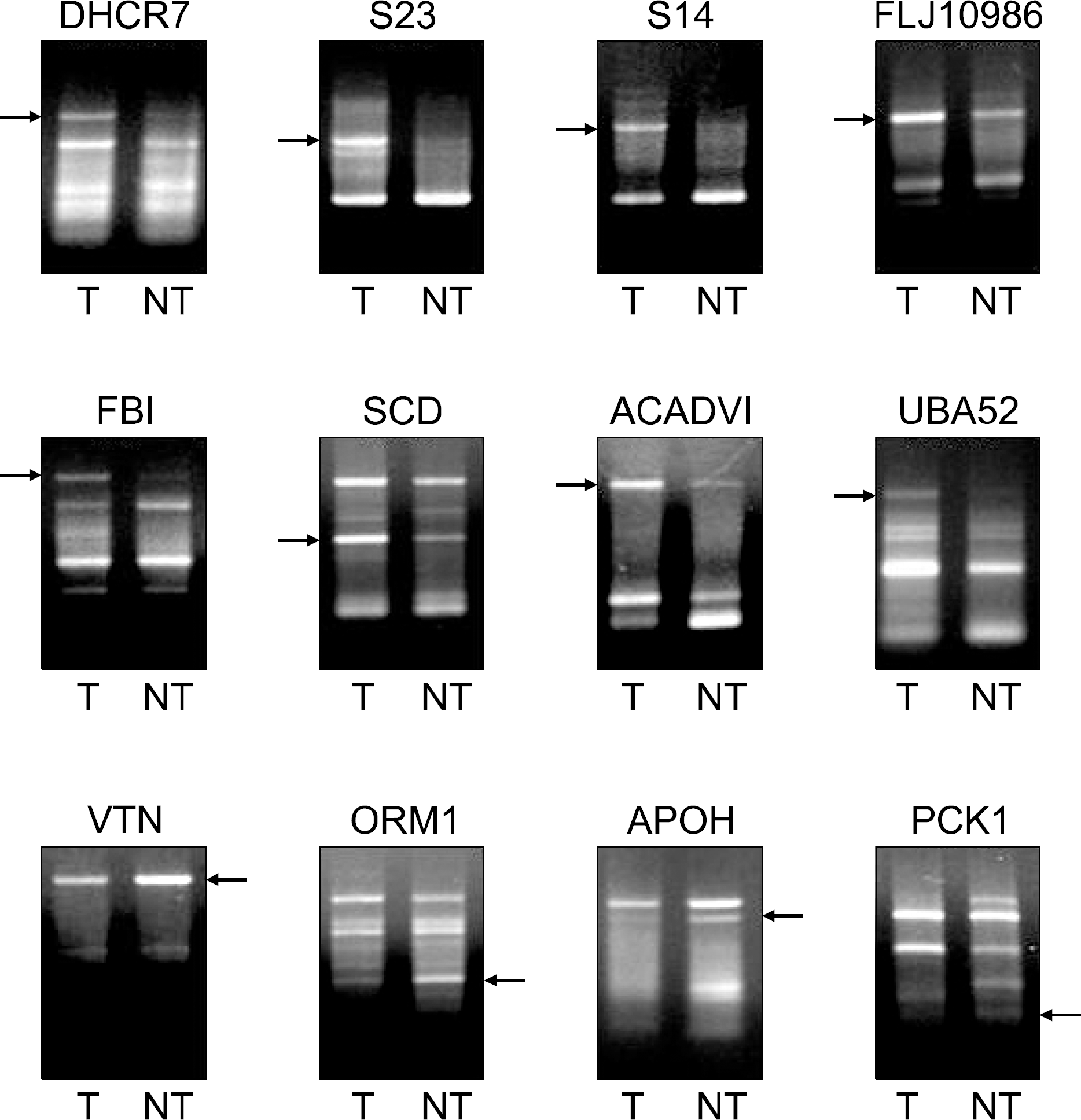 | Fig. 2.Differentially expressed genes (DEGs) between tumor tissue (T) and non-tumor tissue (NT). More than hundreds of DEGs were generated. Among them, 25 DEGs were markedly up-regulated and 10 DEGs were down-regulated in T. These DEG bands were excised from the gel for further cloning and sequencing. |
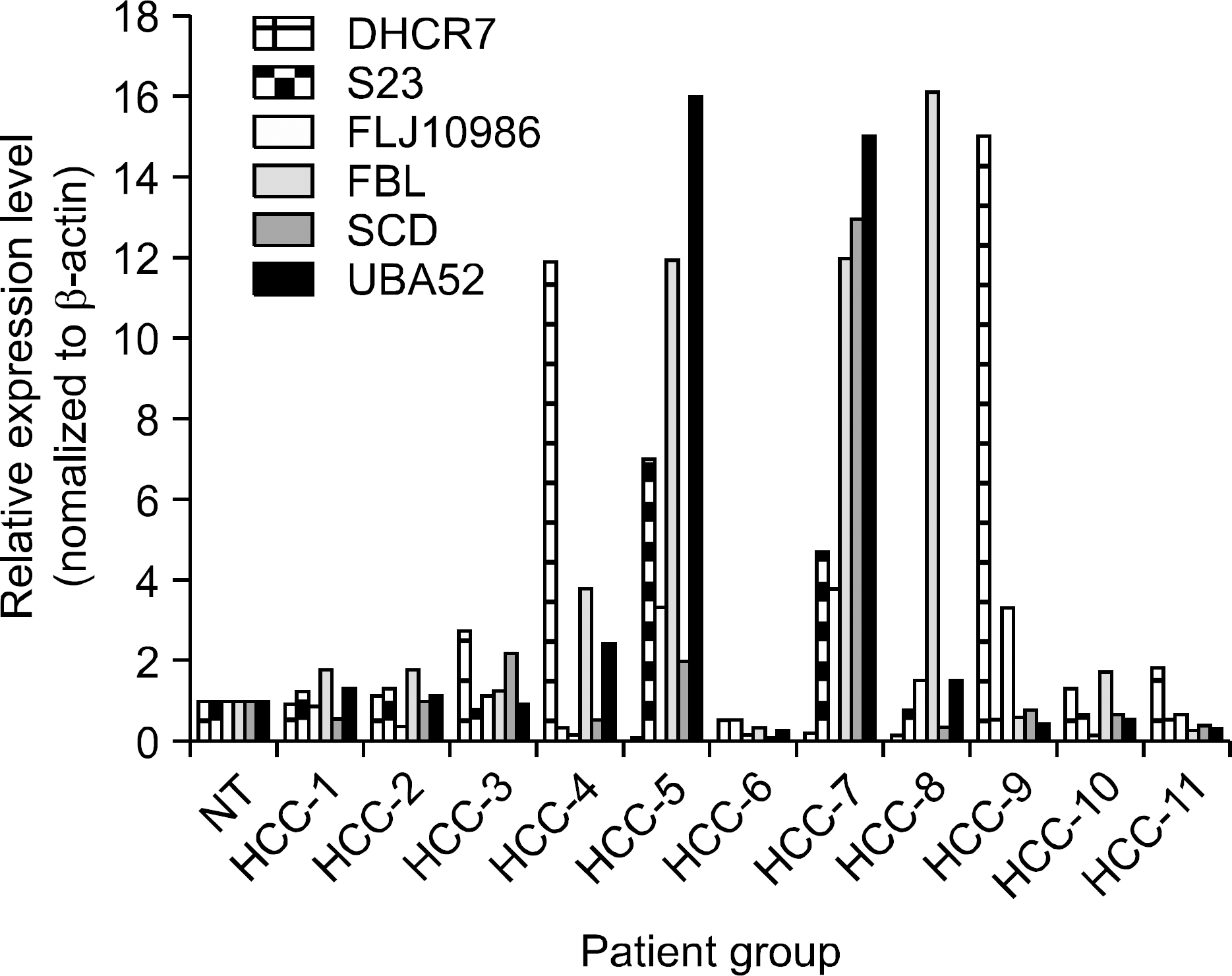 | Fig. 3.Relative expression levels measured by semi-quantitative RT-PCR. In order to validate the observed expression on the up-regulated genes (DHCR7, S23, FLJ10986, FBL, SCD, UBA52), we performed quantitative real-time RT-PCR in 11 HCCs and non-tumor tissue (NT). The obtained mRNA level for FBL was elevated in 8 of 11 in HCCs, and DHCR7, and UBA52 were elevated in 6 of 11 HCCs, respectively. All PCRs were conducted in triplicate and normalized for β-actin mRNA expression. Also, data are presented as the level of expression in each HCCs with respect to the coresponding NT. |
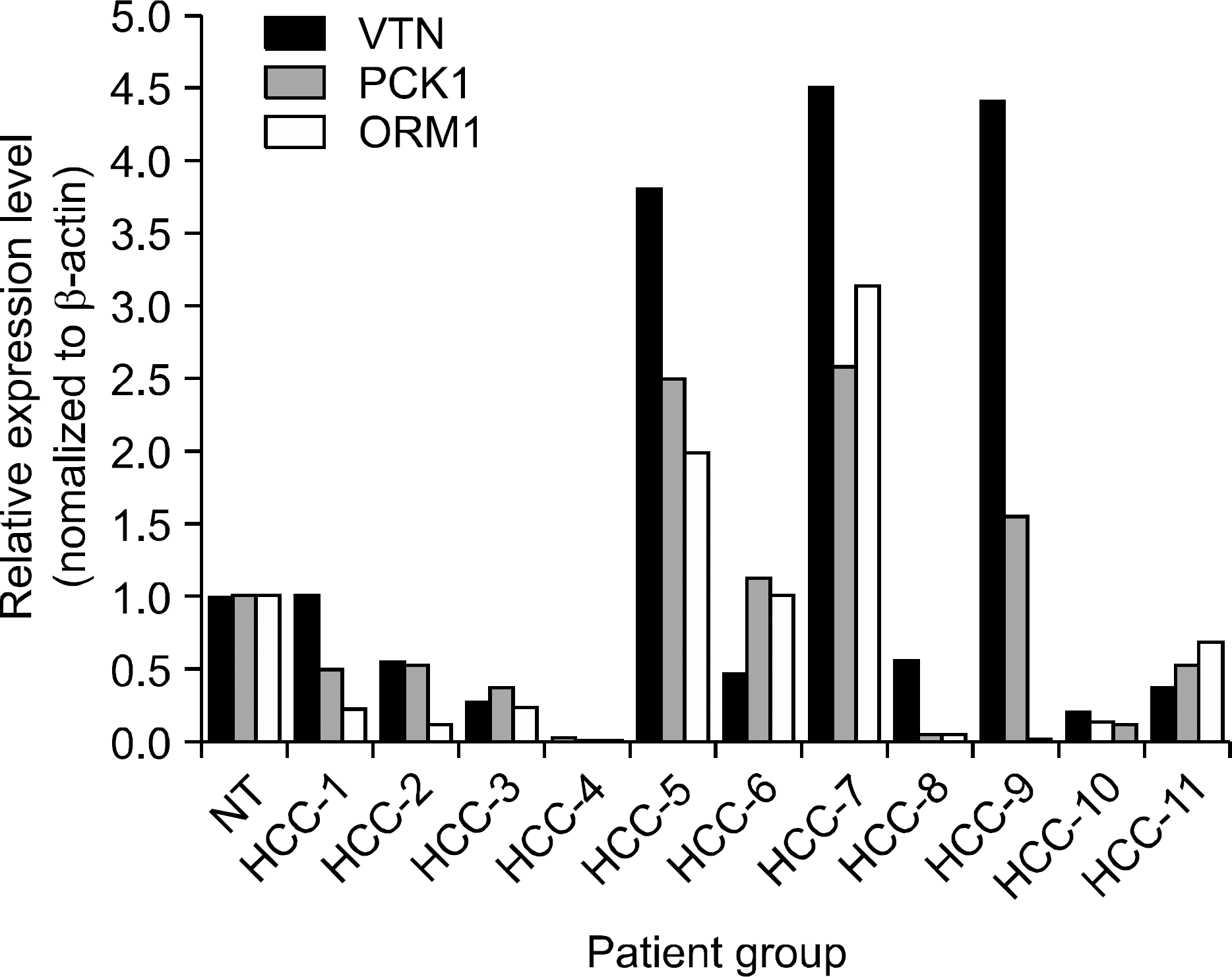 | Fig. 4.Relative expression levels measured by semi-quantitative RT-PCR. In order to validate the observed expression on the down-regulated genes (VTN, PCK1, ORM1), we performed quantitative real-time RT-PCR in 11 HCCs and non-tumor tissue (NT). The obtained mRNA levels for VTN, and ORM1 were on-ly elevated in 3 of 11, and PCK1 was elevated in 4 of 11 in HCCs. All PCRs were conducted in triplicate and normalized for β-actin mRNA expression. Also, data are presented as the level of expression in each HCCs with respect to the corresponding NT. |
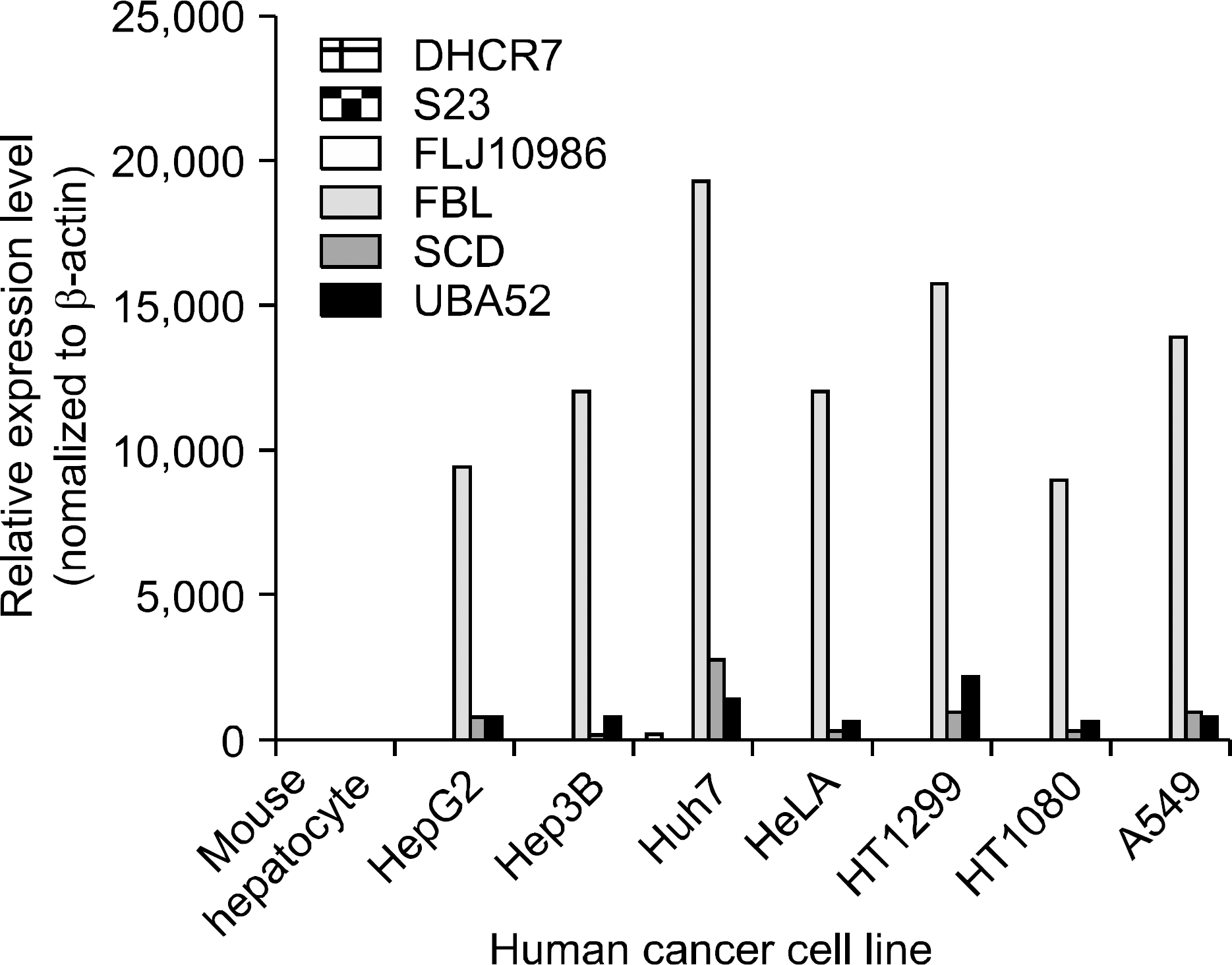 | Fig. 5.Relative expression levels measured by semi-quantitative RT-PCR. In order to validate the observed expression on the up-regulated genes (DHCR7, S23, FLJ10986, FBL, SCD, UBA52), we performed quantitative real-time RT-PCR in different human cancer cell lines. The obtained mRNA levels for DHCR7, S23, FLJ10986, FBL, SCD, and UBA52 were elevated in 7 of 7 human cancer cell lines. All PCRs were conducted in triplicate and normalized for β-actin mRNA expression. Also, data are presented as the level of expression in each human cancer cell lines with respect to the corresponding mouse hepatocyte. |
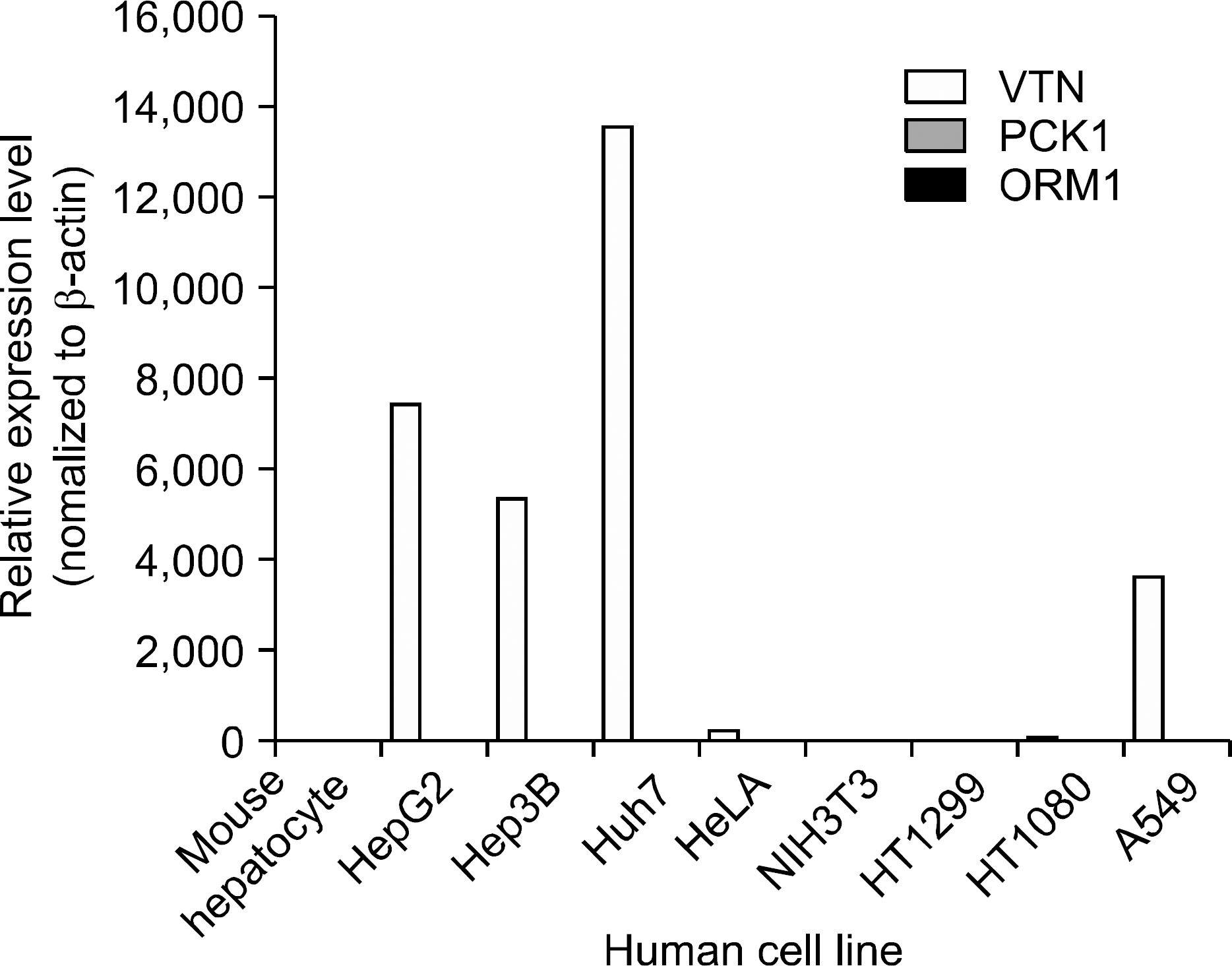 | Fig. 6.Relative expression levels measured by semi-quantitative RT-PCR. In order to validate the observed expression on the down-regulated genes (VTN, PCK1, ORM1), we performed quantitative real-time RT-PCR in different human cancer cell lines. The obtained mRNA levels for VTN, and ORM1 were elevated in 5 of 7 and PCK1 was elevated in 4 of 7 human cancer cell lines. All PCRs were conducted in triplicate and normalized for β-actin mRNA expression. Also, data are presented as the level of expression in each human cancer cell lines with respect to the corresponding mouse hepatocyte. |
Table 1.
The Specific Primer of Genes Identified by Real Time RT-PCR
Table 2.
Up-regulated Genes in Tumor Tissues Compared with Non-tumor Tissues by DDRT-PCR
Table 3.
Down-regulated Genes in Tumor Tissues Compared to Non-tumor Tissues by DDRT-PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download