Abstract
High-grade prostatic intraepithelial neoplasia (HGPIN) has been established as a precursor to prostatic adenocarcinoma. HGPIN shares many morphological, genetic, and molecular signatures with prostate cancer. Its predictive value for the development of future adenocarcinoma during the prostate-specific antigen screening era has decreased, mostly owing to the increase in prostate biopsy cores. Nevertheless, a literature review supports that large-volume HGPIN and multiple cores of involvement at the initial biopsy should prompt a repeat biopsy of the prostate within 1 year. No treatment is recommended for HGPIN to slow its progression to cancer.
The era of prostate-specific antigen (PSA) screening has resulted in an increase in the number of prostate biopsies being performed and as a result an increased detection of prostatic intraepithelial neoplasia (PIN) on microscopic examination [1]. The specific diagnosis of PIN had historically created some debate and uncertainty in terms of its clinical significance and relationship to prostate cancer. However, more recent studies confirm that high-grade PIN (HGPIN) shares a clinical, morphological, and genetic background with prostate cancer. In the current report, we review the recent literature pertaining to this entity and the clinical recommendations once a diagnosis of HGPIN is established.
PIN, which was initially referred to as "intraductal dysplasia," was first described to be a direct biological precursor to prostatic adenocarcinoma by McNeal and Bostwick [2] in 1986. Although in the initial description of PIN the classification included three different grades of dysplasia, at the present time, only HGPIN is reported by pathologists. This is mostly the result of the poor reproducibility among pathologists of distinguishing between low-grade PIN and benign prostate tissue [1,3]. Numerous studies since the initial description have confirmed HGPIN as an accepted precursor to some prostatic adenocarcinomas, and as such it has become a clinically important finding on prostate biopsy in terms of possessing high predictive value for cancer [4]. The estimated time frame to disease progression after HGPIN findings has been reported to be between 29 and 36 months [5,6]. In addition, an increase in the size and number of HGPIN foci has been associated with both prostate adenocarcinoma and its multifocality [7-13]. HGPIN and carcinoma tend to preferentially involve the peripheral zone of the prostate [8,9], and recently many biomarkers and molecular changes such as TMPRSS2-ERG gene fusion have been described in both entities (as discussed further below) [14,15].
The population incidence of HGPIN seems to parallel that of prostate adenocarcinoma. Previous autopsy studies revealed that HGPIN had a low prevalence in men during their third decade of life (7% in African Americans vs. 8% in Caucasians) and progressively increased with advancing age (91% in African Americans vs. 67% in Caucasians) [16]. Sakr et al. [16,17] also observed a higher prevalence of HGPIN in African Americans than in age-matched Caucasian men, similar to prostate cancer. However, the reported incidence of HGPIN in men participating in PSA screening after needle biopsy varies widely according to different series, ranging from 0 to 25% with a mean incidence of 7.7% [4,10,18,19]. Once again, these diverse findings may be explained by the subjectivity of individual pathologists concerning what constitutes HGPIN and possibly by the technical aspects of tissue preparation [18].
The incidence of HGPIN in Asian men is similar to that in Western men. In a study of Korean men undergoing radical cystoprostatectomy, 21% of the men who had no prostate cancer in the final pathology were actually discovered to have HGPIN. As in studies of other races, HGPIN did not correlate with PSA, tumor volume, stage, grade, surgical margins, or lymphovascular invasion [20]. Han et al. [20] also found that both Western and Korean men have high rates of PIN associated with prostate cancer in prostatectomy specimens. In a study from Singapore consisting of 48 Chinese, 3 Malays, 2 Indians, and 3 other men with isolated HGPIN, 24% of those who underwent repeat biopsies had prostate cancer. The authors also observed that most cases of HGPIN affected only one core (79%), with 18% and 4% of cases affecting two and three cores, respectively [21].
Historically, isolated HGPIN on the initial core biopsy was reported to be associated with a very high risk of prostate cancer on the repeat biopsy, up to 80% in some series [5,18,22-24]. In the contemporary era, this risk seems to have decreased to approximately 24%, which is slightly higher than the 19% found after repeat biopsy for a benign diagnosis [5,18]. This decrease in the incidence of cancer following a diagnosis of HGPIN is likely related to the increase in the number of needle core biopsies taken, which has improved the accuracy of the initial biopsy. As one would expect with a pre-malignant lesion, the rate of cancer detection on repeat biopsy has been strongly correlated with an increasing number of cores (≥2) containing HGPIN in the initial biopsy [7,12,13]. As a result, this has led to the recommendation of repeat prostate biopsy within 1 year of such findings. Interestingly, neither the initial PSA level nor PSA velocity at the time of HGPIN discovery correlates with the risk of cancer on repeat biopsy [12,13]. Likewise, digital rectal examination, transrectal ultrasound, and other imaging studies have not been shown to be useful in detecting HGPIN [12,13].
More than 50 genetic and molecular abnormalities have been associated with HGPIN. At least 10 of these changes are reported in both HGPIN and prostate cancer [25-33]. In addition, more than 36 genetic and molecular alterations are intermediate between normal prostate and prostate cancer [25]. This has led to multiple studies making the conclusion that "High-grade PIN is the most likely precursor of prostatic adenocarcinoma, according to virtually all available evidence" [10,34,35]. Two excellent reviews that list in great detail the known genetic and molecular changes in HGPIN were published by Bostwick and Qian [25] and Mosquera et al. [33], and readers are encouraged to read them.
The TMPRSS2-ERG gene fusion has been found in 30 to 79% of prostate cancers [36] and is associated with aggressive disease in some studies [37,38] but not in others [39]. This fusion has also been discovered in 16 to 19% of HGPIN lesions in patients with prostate cancer [15,33]. Because the cancer had the same fusion pattern as the HGPIN in all cases, it has been postulated that the HGPIN may be the premalignant lesion from which the cancer arose [33].
Multiple mechanisms leading to the dysregulation of cell proliferation and survival have been observed to be present in HGPIN. For example, overexpression of Prostate Tumor Overexpressed-1 contributes to cell proliferation in prostate cancer. In HGPIN, it was shown to be an independent predictor of prostate cancer on repeat biopsy [40]. In addition, up-regulation of the mammalian target of rapamycin (mTOR) pathway promotes prostate cancer. A protein known as 14-3-3σ, which regulates the mTOR pathway, shows progressively increasing levels of expression from HGPIN through Gleason score 6, Gleason score 7, and high-grade prostate cancer [41]. The RER+ phenotype leads to the inactivation of many tumor suppressor genes. In one study, the RER+ phenotype was found in 4% of non-cancerous prostate tissues, 16% of PIN lesions, and 42% of prostate cancer lesions [27]. The same study found microsatellite instability in 12%, 35%, and 53% of normal, PIN, and cancerous tissues. Fatty acid synthetase (FAS) expression, which is thought to be an early event in malignant transformation, has been shown to be overexpressed in HGPIN and prostate cancer, but no FAS expression was observed in normal tissue [42]. Likewise, P53 mutations were detected in 14% of PIN and 25% of prostate cancer lesions in one study [43]. Bcl-2, a proto-oncogene that inhibits apoptosis, has been expressed in many prostate cancers, and has also been reported by Baltaci et al. [28] to be present in both low-grade PIN and HGPIN after immunohistochemical evaluation.
Abnormalities in chromosomes, chromatin structure, and DNA processing enzymes have also been described in HGPIN. More than half of HGPIN lesions may have chromosomal anomalies, including gains of chromosomes (decreasing order of frequency) 8, 10, 7, 12, and Y [26,29]. One study discovered loss of heterozygosity on chromosome 8p12-21 in 63% of PIN lesions in patients with prostate cancer [26]. Telomerase, which can contribute to cancer cell immortalization, is active is some HGPIN foci [32]. DNA topoisomerase II-alpha staining by immunohistochemistry is intermediate between benign tissue and prostate cancer [30]. HGPIN also displays epigenetic changes, including hypermethylation [44].
HGPIN is characterized by a proliferation of secretory cells with significant cytologic atypia within the prostate glands and acini. The secretory cells are enlarged with an increased nuclear/cytoplasmic ratio and prominent nucleoli (Fig. 1). The cytoplasm of the HGPIN cells tends to stain positively for α-methylacyl-CoA. Most of these features are shared by invasive prostate cancer [10,25,45]. However, in contrast to prostatic adenocarcinoma in which the basal cells are absent, in HGPIN the basal cell layer is retained although it is often discontinuous on hematoxylin and eosin stain (Fig. 2) [45]. Immunohistochemical staining with antibodies to high molecular weight cytokeratins or nuclear p63 may help in the diagnosis because the presence of basal cells is easily demonstrated [10,19].
Perhaps the most important question about HGPIN is whether its presence signals a higher risk of developing prostate cancer. For men who do not have cancer on prostate biopsy, are those with HGPIN more likely to be diagnosed with prostate cancer in the future than are those with only benign tissue on the initial biopsy? Historically, the answer was "yes." Prior to the era of intense PSA screening and before the implementation of extended prostate biopsy schemes, HGPIN indicated up to an 80% probability of finding prostate cancer on additional biopsies [22]. At the time, it was astutely observed that "The finding of high-grade PIN on needle biopsy often represents a sampling problem with carcinoma nearby" [22]. Because the number of cores routinely sampled at prostate biopsy has increased, the predictive value of HGPIN for prostate cancer has decreased. In studies published before 2000, the prostate cancer detection rate was 36% on follow-up biopsy, but after 2000, this rate dropped to 21% [47]. Another review of studies published after 2000 found the risk of cancer on subsequent biopsy to be 5% lower for patients with HGPIN than for those with benign disease [18]. Two reviews by Bostwick et al. tabulate the details of multiple studies of the cancer detection rate in patients with HGPIN [25,48].
Several studies have shown that taking at least 8 cores on the initial biopsy leads to a low prostate cancer detection rate on repeat biopsy [49,50]. In men diagnosed with HGPIN during initial extended biopsies (at least 10 cores), taking more cores (at least 20) on repeat biopsies was an independent predictor of prostate cancer detection, after multivariate adjustment for age, PSA, days from initial biopsy, DRE status, and multifocal PIN [51]. In-office 24-core saturation biopsy detects prostate cancer in 18% of men with HGPIN detected on a prior biopsy [52].
Multifocal HGPIN on biopsy predicts a higher rate of prostate cancer on repeat biopsy than does a single focus of HGPIN [13,53,54]. The evidence for this finding has been noted in multiple studies, and multifocality has been established as a consistent predictor. Roscigno et al. [53] incorporated multifocality along with PSA level, age, and ≤ 12-core initial biopsy into a nomogram that was 72% accurate in predicting prostate cancer on repeat biopsy. The current data on the prognostic value of single versus multiple cores of HGPIN are summarized in Table 1. Studies on the rate of subsequent cancer diagnosis in men with HGPIN on limited vs. extended vs. saturation biopsy schemes are summarized in Table 2.
The management of patients with HGPIN depends largely on the interpretation of the data presented above. Guidelines from the European Association of Urology state that "HGPIN as an isolated finding is no longer considered an indication for re-biopsy" unless it occurs multifocally [55]. The National Comprehensive Cancer Network acknowledges the need for an extended biopsy scheme in their guidelines that "recommend a repeat biopsy using an extended pattern including transition zone if non-focal HGPIN is found on a sextant biopsy. If extended biopsies were used initially, a delayed strategy (re-biopsy 1 year after initial biopsy) may be considered" [56].
Despite the similar rates of cancer detection in men with monofocal HGPIN and benign biopsies, some sources continue to recommend repeat biopsy at 1 year for such findings [48]. However, we feel that based on the currently available data, men with monofocal HGPIN do not need a scheduled repeat biopsy, but that the decision for repeat biopsy should be based on standard clinical factors (rising PSA, new nodule on DRE, etc). Others also espouse this approach [18,57]. We also feel that the evidence supports repeat biopsy for multifocal HGPIN and for patients who had fewer than 10 cores taken on the initial biopsy. Given the low correlation between HGPIN and prostate cancer, nearly all sources recommend against radical prostatectomy, radiation, or androgen deprivation therapy for isolated HGPIN.
A number of studies have investigated possible therapies to lower the incidence of HGPIN or to decrease the rate of progression from HGPIN to prostate cancer [48]. The Prostate Cancer Prevention Trial demonstrated that 7 years of treatment with daily finasteride decreased the incidence of HGPIN from 7.1 to 6.0% [58]. Similarly, in the REDUCE trial, dutasteride reduced the incidence of HGPIN from 6.0 to 3.7% [59]. Green tea catechins for 1 year in men with HGPIN reduced the incidence of cancer from 30 to 3% [60]. Soy, vitamin E, and selenium did not slow the rate of progression of HGPIN to prostate cancer in a randomized double-blind trial in 303 men [61]. We do not routinely recommend any of these therapies to men with HGPIN.
HGPIN is a relatively common finding in prostate biopsies, and it has been established as a precursor to prostate adenocarcinoma. Its predictive value of increased risk of developing future cancer is low when identified in isolation or in low volume. However, a repeat biopsy should be strongly considered if the initial biopsy contains less than 10 cores or if the HGPIN is present in large volume and within multiple cores.
Figures and Tables
FIG. 1
Prostatic gland with HGPIN (right) and normal prostatic gland (left). Note the preserved architecture of the gland involved by HGPIN and largely intact basal cell layer (H&E, ×20). HGPIN, high-grade prostatic intraepithelial neoplasia.
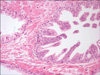
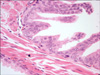
FIG. 2
High-power view of prostatic gland with HGPIN showing enlarged secretory cells with increased nuclear/cytoplasmic ratio and prominent nucleoli. Note the partially disrupted basal cell layer (H&E, ×40). HGPIN, high-grade prostatic intraepithelial neoplasia.
References
1. Epstein JI, Grignon DJ, Humphrey PA, McNeal JE, Sesterhenn IA, Troncoso P, et al. Interobserver reproducibility in the diagnosis of prostatic intraepithelial neoplasia. Am J Surg Pathol. 1995. 19:873–886.
2. McNeal JE, Bostwick DG. Intraductal dysplasia: a premalignant lesion of the prostate. Hum Pathol. 1986. 17:64–71.
3. Allam CK, Bostwick DG, Hayes JA, Upton MP, Wade GG, Domanowski GF, et al. Interobserver variability in the diagnosis of high-grade prostatic intraepithelial neoplasia and adenocarcinoma. Mod Pathol. 1996. 9:742–751.
4. Bostwick DG, Cheng L. Precursors of prostate cancer. Histopathology. 2012. 60:4–27.
5. Guzzo TJ, Kutikov A, Canter DJ, Tomaszewski JE, Magerfleish L, VanArsdalen K, et al. The clinical and pathological history of prostate cancer progression in men with a prior history of high grade prostatic intraepithelial neoplasia. Can J Urol. 2008. 15:4174–4178.
6. Lefkowitz GK, Taneja SS, Brown J, Melamed J, Lepor H. Followup interval prostate biopsy 3 years after diagnosis of high grade prostatic intraepithelial neoplasia is associated with high likelihood of prostate cancer, independent of change in prostate specific antigen levels. J Urol. 2002. 168:1415–1418.
7. Antonelli A, Tardanico R, Giovanessi L, Pesenti N, Gatti L, Zambolin T, et al. Predicting prostate cancer at rebiopsies in patients with high-grade prostatic intraepithelial neoplasia: a study on 546 patients. Prostate Cancer Prostatic Dis. 2011. 14:173–176.
8. Bostwick DG. Prospective origins of prostate carcinoma. Prostatic intraepithelial neoplasia and atypical adenomatous hyperplasia. Cancer. 1996. 78:330–336.
9. Häggman MJ, Macoska JA, Wojno KJ, Oesterling JE. The relationship between prostatic intraepithelial neoplasia and prostate cancer: critical issues. J Urol. 1997. 158:12–22.
10. Montironi R, Mazzucchelli R, Lopez-Beltran A, Cheng L, Scarpelli M. Mechanisms of disease: high-grade prostatic intraepithelial neoplasia and other proposed preneoplastic lesions in the prostate. Nat Clin Pract Urol. 2007. 4:321–332.
11. Abdel-Khalek M, El-Baz M, Ibrahiem el-H. Predictors of prostate cancer on extended biopsy in patients with high-grade prostatic intraepithelial neoplasia: a multivariate analysis model. BJU Int. 2004. 94:528–533.
12. Roscigno M, Scattoni V, Freschi M, Raber M, Colombo R, Bertini R, et al. Monofocal and plurifocal high-grade prostatic intraepithelial neoplasia on extended prostate biopsies: factors predicting cancer detection on extended repeat biopsy. Urology. 2004. 63:1105–1110.
13. Kronz JD, Allan CH, Shaikh AA, Epstein JI. Predicting cancer following a diagnosis of high-grade prostatic intraepithelial neoplasia on needle biopsy: data on men with more than one follow-up biopsy. Am J Surg Pathol. 2001. 25:1079–1085.
14. Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jerónimo C, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006. 8:826–832.
15. Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007. 31:882–888.
16. Sakr WA, Grignon DJ, Haas GP, Heilbrun LK, Pontes JE, Crissman JD. Age and racial distribution of prostatic intraepithelial neoplasia. Eur Urol. 1996. 30:138–144.
17. Sakr WA, Grignon DJ, Haas GP. Pathology of premalignant lesions and carcinoma of the prostate in African-American men. Semin Urol Oncol. 1998. 16:214–220.
18. Epstein JI, Herawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol. 2006. 175:820–834.
19. Zynger DL, Yang X. High-grade prostatic intraepithelial neoplasia of the prostate: the precursor lesion of prostate cancer. Int J Clin Exp Pathol. 2009. 2:327–338.
20. Han KS, Jeong IG, Joung JY, Yang SO, Chung J, Seo HK, et al. Prevalence of high-grade prostatic intraepithelial neoplasia in prostate gland of Korean men: comparisons between radical prostatectomy and cystoprostatectomy. Urology. 2007. 70:1100–1103.
21. Tan PH, Tan HW, Tan Y, Lim CN, Cheng C, Epstein JI, et al. Is high-grade prostatic intraepithelial neoplasia on needle biopsy different in an Asian population: a clinicopathologic study performed in Singapore. Urology. 2006. 68:800–803.
22. Weinstein MH, Epstein JI. Significance of high-grade prostatic intraepithelial neoplasia on needle biopsy. Hum Pathol. 1993. 24:624–629.
23. Keetch DW, Humphrey P, Stahl D, Smith DS, Catalona WJ. Morphometric analysis and clinical followup of isolated prostatic intraepithelial neoplasia in needle biopsy of the prostate. J Urol. 1995. 154:347–351.
24. Park S, Shinohara K, Grossfeld GD, Carroll PR. Prostate cancer detection in men with prior high grade prostatic intraepithelial neoplasia or atypical prostate biopsy. J Urol. 2001. 165:1409–1414.
25. Bostwick DG, Qian J. High-grade prostatic intraepithelial neoplasia. Mod Pathol. 2004. 17:360–379.
26. Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, et al. Allelic loss on chromosome 8p12-21 in microdissected prostatic intraepithelial neoplasia. Cancer Res. 1995. 55:2959–2962.
27. Miet SM, Neyra M, Jaques R, Dubernard P, Revol AA, Marçais C. RER(+) phenotype in prostate intra-epithelial neoplasia associated with human prostate-carcinoma development. Int J Cancer. 1999. 82:635–639.
28. Baltaci S, Orhan D, Ozer G, Tolunay O, Gögõüs O. Bcl-2 proto-oncogene expression in low- and high-grade prostatic intraepithelial neoplasia. BJU Int. 2000. 85:155–159.
29. Qian J, Bostwick DG, Takahashi S, Borell TJ, Herath JF, Lieber MM, et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Res. 1995. 55:5408–5414.
30. Willman JH, Holden JA. Immunohistochemical staining for DNA topoisomerase II-alpha in benign, premalignant, and malignant lesions of the prostate. Prostate. 2000. 42:280–286.
31. Ge K, Minhas F, Duhadaway J, Mao NC, Wilson D, Buccafusca R, et al. Loss of heterozygosity and tumor suppressor activity of Bin1 in prostate carcinoma. Int J Cancer. 2000. 86:155–161.
32. Koeneman KS, Pan CX, Jin JK, Pyle JM 3rd, Flanigan RC, Shankey TV, et al. Telomerase activity, telomere length, and DNA ploidy in prostatic intraepithelial neoplasia (PIN). J Urol. 1998. 160:1533–1539.
33. Mosquera JM, Perner S, Genega EM, Sanda M, Hofer MD, Mertz KD, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008. 14:3380–3385.
34. Bostwick DG, Pacelli A, Lopez-Beltran A. Molecular biology of prostatic intraepithelial neoplasia. Prostate. 1996. 29:117–134.
35. Montironi R, Mazzucchelli R, Lopez-Beltran A, Scarpelli M, Cheng L. Prostatic intraepithelial neoplasia: its morphological and molecular diagnosis and clinical significance. BJU Int. 2011. 108:1394–1401.
36. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005. 310:644–648.
37. Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008. 27:253–263.
38. Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006. 66:8337–8341.
39. Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009. 69:1400–1406.
40. Morote J, Fernández S, Alaña L, Iglesias C, Planas J, Reventós J, et al. PTOV1 expression predicts prostate cancer in men with isolated high-grade prostatic intraepithelial neoplasia in needle biopsy. Clin Cancer Res. 2008. 14:2617–2622.
41. Evren S, Dermen A, Lockwood G, Fleshner N, Sweet J. mTOR-RAPTOR and 14-3-3sigma immunohistochemical expression in high grade prostatic intraepithelial neoplasia and prostatic adenocarcinomas: a tissue microarray study. J Clin Pathol. 2011. 64:683–688.
42. Swinnen JV, Roskams T, Joniau S, Van Poppel H, Oyen R, Baert L, et al. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int J Cancer. 2002. 98:19–22.
43. Yasunaga Y, Shin M, Fujita MQ, Nonomura N, Miki T, Okuyama A, et al. Different patterns of p53 mutations in prostatic intraepithelial neoplasia and concurrent carcinoma: analysis of microdissected specimens. Lab Invest. 1998. 78:1275–1279.
44. Brooks JD, Weinstein M, Lin X, Sun Y, Pin SS, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomarkers Prev. 1998. 7:531–536.
45. Bostwick DG, Brawer MK. Prostatic intra-epithelial neoplasia and early invasion in prostate cancer. Cancer. 1987. 59:788–794.
46. Bostwick DG, Amin MB, Dundore P, Marsh W, Schultz DS. Architectural patterns of high-grade prostatic intraepithelial neoplasia. Hum Pathol. 1993. 24:298–310.
47. Schlesinger C, Bostwick DG, Iczkowski KA. High-grade prostatic intraepithelial neoplasia and atypical small acinar proliferation: predictive value for cancer in current practice. Am J Surg Pathol. 2005. 29:1201–1207.
48. Bostwick DG, Cheng L. Precursors of prostate cancer. Histopathology. 2012. 60:4–27.
49. Herawi M, Kahane H, Cavallo C, Epstein JI. Risk of prostate cancer on first re-biopsy within 1 year following a diagnosis of high grade prostatic intraepithelial neoplasia is related to the number of cores sampled. J Urol. 2006. 175:121–124.
50. Eskicorapci SY, Guliyev F, Islamoglu E, Ergen A, Ozen H. The effect of prior biopsy scheme on prostate cancer detection for repeat biopsy population: results of the 14-core prostate biopsy technique. Int Urol Nephrol. 2007. 39:189–195.
51. Lee MC, Moussa AS, Zaytoun O, Yu C, Jones JS. Using a saturation biopsy scheme increases cancer detection during repeat biopsy in men with high-grade prostatic intra-epithelial neoplasia. Urology. 2011. 78:1115–1119.
52. Rabets JC, Jones JS, Patel A, Zippe CD. Prostate cancer detection with office based saturation biopsy in a repeat biopsy population. J Urol. 2004. 172:94–97.
53. Roscigno M, Scattoni V, Freschi M, Abdollah F, Maccagnano C, Galosi A, et al. Diagnosis of isolated high-grade prostatic intra-epithelial neoplasia: proposal of a nomogram for the prediction of cancer detection at saturation re-biopsy. BJU Int. 2011. Epub ahead of print.
54. Akhavan A, Keith JD, Bastacky SI, Cai C, Wang Y, Nelson JB. The proportion of cores with high-grade prostatic intraepithelial neoplasia on extended-pattern needle biopsy is significantly associated with prostate cancer on site-directed repeat biopsy. BJU Int. 2007. 99:765–769.
55. Heidenreich A, Bellmunt J, Bolla M, Joniau S, Mason M, Matveev V, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011. 59:61–71.
56. Kawachi MH, Bahnson RR, Barry M, Busby JE, Carroll PR, Carter HB, et al. NCCN clinical practice guidelines in oncology: prostate cancer early detection. J Natl Compr Canc Netw. 2010. 8:240–262.
57. Gallo F, Chiono L, Gastaldi E, Venturino E, Giberti C. Prognostic significance of high-grade prostatic intraepithelial neoplasia (HGPIN): risk of prostatic cancer on repeat biopsies. Urology. 2008. 72:628–632.
58. Thompson IM, Lucia MS, Redman MW, Darke A, La Rosa FG, Parnes HL, et al. Finasteride decreases the risk of prostatic intraepithelial neoplasia. J Urol. 2007. 178:107–109.
59. Andriole GL, Bostwick DG, Brawley OW, Gomella LG, Marberger M, Montorsi F, et al. Effect of dutasteride on the risk of prostate cancer. N Engl J Med. 2010. 362:1192–1202.
60. Bettuzzi S, Brausi M, Rizzi F, Castagnetti G, Peracchia G, Corti A. Chemoprevention of human prostate cancer by oral administration of green tea catechins in volunteers with high-grade prostate intraepithelial neoplasia: a preliminary report from a one-year proof-of-principle study. Cancer Res. 2006. 66:1234–1240.
61. Fleshner NE, Kapusta L, Donnelly B, Tanguay S, Chin J, Hersey K, et al. Progression from high-grade prostatic intraepithelial neoplasia to cancer: a randomized trial of combination vitamin-E, soy, and selenium. J Clin Oncol. 2011. 29:2386–2390.
62. Lefkowitz GK, Sidhu GS, Torre P, Lepor H, Taneja SS. Is repeat prostate biopsy for high-grade prostatic intraepithelial neoplasia necessary after routine 12-core sampling? Urology. 2001. 58:999–1003.
63. Moore CK, Karikehalli S, Nazeer T, Fisher HA, Kaufman RP Jr, Mian BM. Prognostic significance of high grade prostatic intraepithelial neoplasia and atypical small acinar proliferation in the contemporary era. J Urol. 2005. 173:70–72.
64. Schoenfield L, Jones JS, Zippe CD, Reuther AM, Klein E, Zhou M, et al. The incidence of high-grade prostatic intraepithelial neoplasia and atypical glands suspicious for carcinoma on first-time saturation needle biopsy, and the subsequent risk of cancer. BJU Int. 2007. 99:770–774.
65. De Nunzio C, Trucchi A, Miano R, Stoppacciaro A, Fattahi H, Cicione A, et al. The number of cores positive for high grade prostatic intraepithelial neoplasia on initial biopsy is associated with prostate cancer on second biopsy. J Urol. 2009. 181:1069–1074.
66. Antonelli A, Tardanico R, Giovanessi L, Pesenti N, Gatti L, Zambolin T, et al. Predicting prostate cancer at rebiopsies in patients with high-grade prostatic intraepithelial neoplasia: a study on 546 patients. Prostate Cancer Prostatic Dis. 2011. 14:173–176.
67. Bishara T, Ramnani DM, Epstein JI. High-grade prostatic intraepithelial neoplasia on needle biopsy: risk of cancer on repeat biopsy related to number of involved cores and morphologic pattern. Am J Surg Pathol. 2004. 28:629–633.
68. Netto GJ, Epstein JI. Widespread high-grade prostatic intraepithelial neoplasia on prostatic needle biopsy: a significant likelihood of subsequently diagnosed adenocarcinoma. Am J Surg Pathol. 2006. 30:1184–1188.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download