Abstract
Purpose
The glutathione-S-transferase (GST)P1, GSTM1, and GSTT1 genotypes have been associated with an increased risk of prostate, bladder, and lung cancers. The aim of this study was to investigate the association between the GSTP1, GSTM1, and GSTT1 genotypes and the risk of prostate cancer in Korean men.
Materials and Methods
The study group consisted of 166 patients with histologically confirmed prostate cancer. The control group consisted of 327 healthy, cancer-free individuals. The diagnosis of prostate cancer was made by transrectal ultrasound-guided biopsy. Patients with prostatic adenocarcinoma were divided into organ-confined (≤pT2) and non-organ-confined (≥pT3) subgroups. The histological grades were subdivided according to the Gleason score. The GSTP1, GSTM1, and GSTT1 genotypes were determined by using polymerase chain reaction-based methods. The relationship among GSTP1, GSTM1, and GSTT1 polymorphisms and prostate cancer in a case-control study was investigated.
Results
The frequency of the GSTM1 null genotype in the prostate cancer group (54.2%) was higher than in the control group (odds ratio=1.53, 95% confidence interval=1.20-1.96). The comparison of the GSTP1, GSTM1, and GSTT1 genotypes and cancer prognostic factors, such as staging and grading, showed no statistical significance.
The biotransformation enzymes, glutathione-S-transferases (GSTs), are members of a multigene family; their gene products are phase II enzymes with both catalytic activities, including glutathione conjugation of electrophiles, and noncatalytic functions. The presumed function of these enzymes is to protect tissues against toxic and carcinogenic compounds that enter the body as either food additives or drugs [1]. In addition to their catalytic activities, GSTs are thought to engage metabolites and steroid hormones, which are important determinants in the development of prostate cancer [2,3]. The GSTs are involved in the detoxification of electrophilic compounds (such as carcinogens and cytotoxic drugs) by glutathione conjugation [4,5]. In addition, these enzymes are thought to play a role in the protection of DNA from oxidative damage [6]. GSTP1 inactivation may lead to increased cell vulnerability to oxidative DNA damage and to the accumulation of DNA base adducts, which allows tumors to acquire other relevant genetic alterations during prostate carcinogenesis [7].
GSTM1 detoxifies carcinogenic polycyclic aromatic hydrocarbons, such as the smoke carcinogen benzopyrene, whereas GSTT1 detoxifies smaller reactive hydrocarbons, such as ethylene oxide. The GSTM1 null genotype has a decreased capacity to detoxify certain carcinogens and has been linked with an increased risk for solid tumors [8,9]. Examination of the GSTT1 gene may provide insights into the dangers of exposure to common environmental or dietary agents that produce chromosomal damage. Indeed, persons with the GSTT1 null genotype show a reduced ability to detoxify the metabolites of ethylene oxide [7]. The GSTT1 null genotype has been associated with increased risk for ovarian, bladder, and lung cancers [10-13]. However, other studies have not confirmed the association between the GSTT1 null genotype and cancer. The aim of this study was to investigate the association between the GSTP1, GSTM1, and GSTT1 genotypes and the risk of prostate cancer in Korean men.
The DNA samples were provided by the Biobank of Wonkwang University Hospital, which is a member of the National Biobank of Korea; this Biobank is supported by the Ministry of Health, Welfare and Family Affairs. After approval from the institutional review board and informed consent from the participants, genomic DNA was obtained from 166 patients with prostate cancer and from 327 healthy controls. The healthy controls were selected by having a prostate-specific antigen (PSA) value below 2.5 ng/ml, a normal digital rectal examination, and no hypoechoic lesions in transrectal ultrasonography (TRUS). The mean age in the cancer group was 69.6 years (range, 51-87 years), and the mean age in the healthy controls was 68.3 years (range, 50-86 years) (Table 1). Genomic DNA was extracted from the leukocytes of the peripheral blood by means of a standard phenol-chloroform method or with the use of a Genomic DNA Extraction kit (iNtRON Biotechnology, Korea) according to the manufacturer's directions. The diagnosis of prostate cancer was histologically confirmed by TRUS-guided prostate biopsy. The patients were classified on the basis of tumor stage and grade.
The assay reagents for rs1695 and rs1138272 in the GSTP1 gene were designed by Applied Biosystems (Applied Biosystems, USA). The reagents consisted of a 40x mix of unlabeled polymerase chain reaction (PCR) primer and TaqMan MGB probes (FAM and VIC dye-labeled). The reaction in 10 µl was optimized to work with 0.125 µl 40x reagents, 5 µl 2x TaqMan Genotyping Master mix (Applied Biosystems, USA), and 2 µl (50 ng) of genomic DNA. The PCR conditions were as follows: one cycle at 95℃ for 15 min and 40 cycles at 95℃ for 10 s and 60℃ for 45 s. The PCR was performed in the Rotor-Gene thermal cycler RG6000 (Corbett Research, Australia). The samples were read and analyzed by using the Rotor-Gene 1.7.40 software (Corbett Research, Australia).
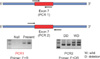
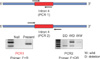
The GSTT1 and GSTM1 genotypes were determined by PCR. The primer pairs used for PCR amplification were 5'-GAACTCCCTGAAAAGCTAAAGC-3' and 5'-GTTGGGCTCAAATATACGGTGG-3' for GSTM1 and 5'-TTCCTTACTGGTCCTCACATCTC-3' and 5'-TCACCGGATCATGGCCAGCA-3' for GSTT1 (Table 2). PCR reactions were carried out for 30 cycles, each including a 10-s denaturation step at 98℃, a 15-s annealing step at 60℃, and a 20-s extension step at 72℃. The PCR program included an initial denaturation time of 5 minutes at 95℃ and an extension time of 10 minutes at 72℃ after the last cycle. The PCR products were separated by electrophoresis by using a 2% agarose gel and were visualized by ethidium bromide staining. The presence of bands at 218 and 459 bp corresponded to intact genomic GSTM1 (Fig. 1) and GSTT1 (Fig. 2), respectively, whereas the absence of the bands implied the null state.
The correlations between the GSTP1, GSTM1, and GSTT1 genotypes and clinico-pathological factors for prostate cancer were analyzed by using SPSS ver. 15.0 (SPSS Inc., Chicago, IL, USA) and analyze software (Dynacom, Yokohama, Japan). The chi-square test was used to calculate the p-values and 95% confidence intervals (95% CIs) for the odds ratios (ORs).
Among the GSTP1 polymorphisms, two single-nucleotide polymorphisms (SNPs; rs1695 and rs1138272) were selected for large sample genotyping on the basis of their locations. The genotype and allele frequencies of rs1695 (g.1375A>G, based on NC_000011.9) were not significantly different between the patients with prostate cancer and the healthy controls (Table 3). The SNP rs1138272 (g.2265C>T), from the NCBI SNP database, was also analyzed for genotype; however, when 96 samples were analyzed, there was only one genotype. These findings suggest that the rs1138272 of GSTP1 might be a very rare polymorphism or a monomorphism in the Korean population.
The frequencies of the GSTM1 null and GSTT1 null genotypes were 38.2% and 49.8% in the control population and 54.2% and 51.2% in the prostate cancer group, respectively. The GSTM1 null genotype was more common in the prostate cancer group than in the control group (OR=1.53, 95% CI=1.20-1.96) (Table 4). No statistically significant correlations were identified between the GSTP1, GSTM1, and GSTT1 genotypes and staging or Gleason score (Table 5, 6, 7).
Prostate cancer is a multifactorial disease that likely involves both environmental and genetic factors. Collectively, most putative environmental and genetic risk factors have not shown a consistent association with prostate cancer risk, and little is known about the interaction of these factors. Prostate cancer risk varies most prominently with age, ethnicity, family history, and diet [14].
Individual differences in the susceptibility to carcinogens play an essential role in the development of sporadic cancer. The biochemical basis for the genetic susceptibility to environmental hazards is related to genetic polymorphisms that normally occur in the general population and involves a series of genes implicated in the metabolic activation or detoxification of environmental genotoxins. Several polymorphic genes encoding enzymes involved in the biotransformation of carcinogens have been studied as possible prostate cancer risk modifiers, including the GST system and the phase I cytochrome P450 (CYP) genes [15].
The GSTs are involved in the metabolism of a wide variety of potential carcinogens. The levels of GST isozymes in normal and tumor tissues are important for several reasons. High levels of GST have been shown to detoxify several chemical carcinogens efficiently and to protect tissues against DNA damage. The presumptive function of GST is to protect tissues against toxic or carcinogenic compounds that enter the body as food components, food additives, or drugs [4-7]. Considerable evidence suggests the existence of various biological defense systems against carcinogenesis. Individuals with homozygous deletions of the GSTM1, GSTT1, and GSTP1 genes lack GST and therefore may be unable to eliminate electrophilic carcinogens efficiently, which may increase the risk of somatic mutations that lead to tumor formation [8-13,16]. The phenotypic absence of GSTM1, GSTT1, and GSTP1 activities is due to homozygous inherited deletions of these genes, which is referred to as the null genotype [17].
Several population-based studies have reported prevalences ranging from 47% to 58% for the GSTM1 deletion genotype and from 13% to 25% for the GSTT1-null genotype among Caucasian Europeans. For GSTP1, the prevalences of Ile/Val heterozygosity and Val/Val homozygosity were found to be between 38% and 45.7% and between 7% and 13%, respectively. Previous studies have shown that the GSTM1 null genotype correlates with increased susceptibility to bladder and prostate cancers, as well as to lung cancer [18,19].
In a Southern European population, an analysis of the frequencies of the 670 alleles indicated that men carrying two B-alleles (GSTM3) have an increased risk for prostate cancer. The polymorphism in GSTM3 may be an important biomarker for prostate cancer risk, especially in the definition of the genetic risk profile of populations of Southern Europe [20]. In Chilean prostate cancer patients, the frequency of the m2 variant allele and GSTM1(-/-) showed statistically significant increases compared with the control group. Chilean people carrying single or combined GSTM1 and CYP1A1 polymorphisms are more susceptible to prostate cancer [21]. In a Brazilian population, the GST and CYP1A1 genotypes were not associated with the susceptibility to prostate cancer or its outcome. Those authors said that they were unable to demonstrate any relationship between genotypes and parameters of aggressiveness at diagnosis or during the follow-up. Also, there was no relationship between the response to radiotherapy and any other outcome [22].
In a Japanese population, the frequency of the GSTM1 null genotype was also slightly higher in prostate cancer patients (49.6%) compared with the control value (42.5%), with an OR of 1.2 (95% CI=0.78-1.99). A GSTT1-positive genotype was thus associated with a 60% higher risk in the prostate cancer group (OR 1.6; 95% CI=0.99-2.51). That study showed a significant relation between the prostate cancer group and the genetic polymorphisms of CYP1A1 alone and in combination with GSTM1 [23]. The GSTP1-313 G polymorphism, and null alleles of GSTM1 and GSTT1, are strong predisposing risk factors for sporadic prostate cancer in North India [24].
In the present study, we investigated the potential link between the GSTM1, GSTT1, and GSTP1 null genotypes and prostate cancer risk. The results of our study showed an association between prostate cancer risk and the presence of the GSTM1 null genotype (Table 3). These results suggest that elevated metabolic activation and decreased levels of detoxification of endogenous or exogenous carcinogens increase DNA adduct formation, thereby increasing the prostate cancer risk [1,19].
The major finding of the present meta-analysis provides support for the association of the genetic polymorphism of GSTM1 (null vs. non-deleted) with the susceptibility to prostate cancer. However, the GSTT1 polymorphism (null vs. nondeleted) and the GSTP1 polymorphism showed no correlation with prostate cancer risk [19]. Although the results of our study show a statistically significant association between the GSTM1 polymorphisms and prostate cancer, the clinical significance of this finding requires further investigation.
There was no significant association between the GSTP1, GSTM1, and GSTT1 genotypes and the clinico-pathologic factors of prostate cancer. To better understand the role of the GSTs and to study their predictive value, tumor prognostic criteria should be examined, such as cancer-specific survival and overall survival.
An increased risk for prostate cancer may be associated with the GSTM1 null genotype in Korean men, but no association was found with the GSTT1 or GSTP1 phenotype. To better understand the role of the GSTs and to study their predictive value, tumor prognostic criteria should be examined, such as cancer-specific survival and overall survival.
Figures and Tables
TABLE 3
Genotype and allele analysis of GSTP1 polymorphisms in patients with prostate cancer and controls

TABLE 5
Correlation of the clinical and pathological features of prostate cancer with GSTP1 genotypes

Notes
References
1. Strange RC, Lear JT, Fryer AA. Glutathione S-transferase polymorphisms: influence on susceptibility to cancer. Chem Biol Interact. 1998. 111-112:351–364.
2. Board P, Harris M, Flanagan J, Langton L, Coggan M. Genetic heterogeneity of the structure and function of GSTT2 and GSTP1. Chem Biol Interact. 1998. 111-112:83–89.
3. Maugard CM, Charrier J, Bignon YJ. Allelic deletion at glutathione S-transferase M1 locus and its association with breast cancer susceptibility. Chem Biol Interact. 1998. 111-112:365–375.
4. Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikula J, Singhal SS, Srivastava SK, et al. Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem. 1994. 224:893–899.
5. Henderson CJ, McLaren AW, Moffat GJ, Bacon EJ, Wolf CR. Pi-class glutathione S-transferase: regulation and function. Chem Biol Interact. 1998. 111-112:69–82.
6. Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, et al. Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis. 1997. 18:1285–1289.
7. Berhane K, Widersten M, Engström A, Kozarich JW, Mannervik B. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proc Natl Acad Sci U S A. 1994. 91:1480–1484.
8. Harada S, Misawa S, Nakamura T, Tanaka N, Ueno E, Nozoe M. Detection of GST1 gene deletion by the polymerase chain reaction and its possible correlation with stomach cancer in Japanese. Hum Genet. 1992. 90:62–64.
9. Nakajima T, Elovaara E, Anttila S, Hirvonen A, Camus AM, Hayes JD, et al. Expression and polymorphism of glutathione S-transferase in human lungs: risk factors in smoking-related lung cancer. Carcinogenesis. 1995. 16:707–711.
10. Abdel-Rahman SZ, Anwar WA, Abdel-Aal WE, Mostafa HM, Au WW. GSTM1 and GSTT1 genes are potential risk modifiers for bladder cancer. Cancer Detect Prev. 1998. 22:129–138.
11. Salagovic J, Kalina I, Stubna J, Habalová M, Hrivnák M, Valanský L, et al. Genetic polymorphism of glutathione S-transferase M1 and T1 as a risk factor in lung and bladder cancers. Neoplasma. 1998. 45:312–317.
12. Hengstler JG, Kett A, Arand M, Oesch-Bartlomowicz B, Oesch F, Pilch H, et al. Glutathione S-transferase T1 and M1 gene defects in ovarian carcinoma. Cancer Lett. 1998. 130:43–48.
13. Katoh T, Inatomi H, Kim H, Yang M, Matsumoto T, Kawamoto T. Effects of glutathione S-transferase (GST) M1 and GSTT1 genotypes on urothelial cancer risk. Cancer Lett. 1998. 132:147–152.
14. Bostwick DG, Burke HB, Djakiew D, Euling S, Ho SM, Landolph J, et al. Human prostate cancer risk factors. Cancer. 2004. 101:10 Suppl. 2371–2490.
15. Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance of familial prostate cancer. Proc Natl Acad Sci U S A. 1992. 89:3367–3371.
16. Nock NL, Liu X, Cicek MS, Li L, Macarie F, Rybicki BA, et al. Polymorphisms in polycyclic aromatic hydrocarbon metabolism and conjugation genes, interactions with smoking and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2006. 15:756–761.
17. Norppa H. Cytogenetic markers of susceptibility: influence of polymorphic carcinogen-metabolizing enzymes. Environ Health Perspect. 1997. 105:Suppl 4. 829–835.
18. Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001. 10:1239–1248.
19. Mo Z, Gao Y, Cao Y, Gao F, Jian L. An updating meta-analysis of the GSTM1, GSTT1, GSTP1 polymorphism and prostate cancer: a HuGE review. Prostate. 2009. 69:662–688.
20. Medeiros R, Vasconcelos A, Costa S, Pinto D, Ferreira P, Lobo F, et al. Metabolic susceptibility genes and prostate cancer risk in a southern European population: the role of glutathione S-transferases GSTM1, GSTM3, and GSTT1 genetic polymorphisms. Prostate. 2004. 58:414–420.
21. Acevedo C, Opazo JL, Huidobro C, Cabezas J, Iturrieta J, Quiñones Sepúlveda L. Positive correlation between single or combined genotypes of CYP1A1 and GSTM1 in relation to porstate cancer in Chilean people. Prostate. 2003. 57:111–117.
22. Lima MM Jr, Oliveira MN, Granja F, Trindade AC, De Castro Santos LE, Ward LS. Lack of association of GSTT1, GSTM1, GSTO1, GSTP1 and CYP1A1 polymorphisms for susceptibility and outcome in Brazilian prostate cancer patients. Folia Biol (Praha). 2008. 54:102–108.
23. Murata M, Watanabe M, Yamanaka M, Kubota Y, Ito H, Nagao M, et al. Genetic polymorphisms in cytochrome P450 (CYP) 1A1, CYP1A2, CYP2E1, glutathione S-transferase (GST) M1 and GSTT1 and susceptibility to prostate cancer in the Japanese population. Cancer Lett. 2001. 165:171–177.
24. Srivastava DS, Mandhani A, Mittal B, Mittal RD. Genetic polymorphism of glutathione S-transferase genes (GSTM1, GSTT1 and GSTP1) and susceptibility to prostate cancer in Northern India. BJU Int. 2005. 95:170–173.




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download