Abstract
Serum is free of cellular components. Because DNA is located in the nuclei or mitochondria of cells, serum could be assumed DNA free. Few previously published case reports to date have used serum for DNA typing. Here, we report on human genotyping via short tandem repeat (STR) analysis using serum as a sample, and discuss problems involved in the process.
Go to : 
REFERENCES
1. Takayama T, Yamada S, Watanabe Y, et al. Origin of DNA in human serum and usefulness of serum as a material for DNA typing. Leg Med (Tokyo). 2001; 3:109–13.

2. Lo YM, Tein MSC, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998; 62:768–75.

3. Honda H, Miharu N, Ohashi Y, et al. Fetal gender determination in early pregnancy through qualitative and quantitative analysis of fetal DNA in maternal serum. Hum Genet. 2002; 110:75–9.

4. Bischoff FZ, Sinacori MK, Dang DD, et al. Cell-free fetal D-NA and intact fetal cells in maternal blood circulation: implications for first and second trimester non-invasive prenatal diagnosis. Hum Reprod Update. 2002; 8:493–500.

5. Allen RW, Pritchard JK. Resolution of a serum sample mix-up through the use of short tandem repeat DNA typing. Transfusion. 2004; 44:1750–4.

6. Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350(9076):485–7.

7. Ravard-Goulvestre C, Crainic K, Guillon F, et al. Successful extraction of human genomic DNA from serum and its application to forensic identification. J Forensic Sci. 2004; 49:60–3.

8. Emanuel SL, Pestka S. Amplification of specific gene products from human serum. Genetic Analysis: Biomolecular Engineering. 1993; 10:144–6.

9. Butler J.M.Fundamentals of forensic DNA typing. Access Online via Elsevier. 2009. 330–39.
Go to : 
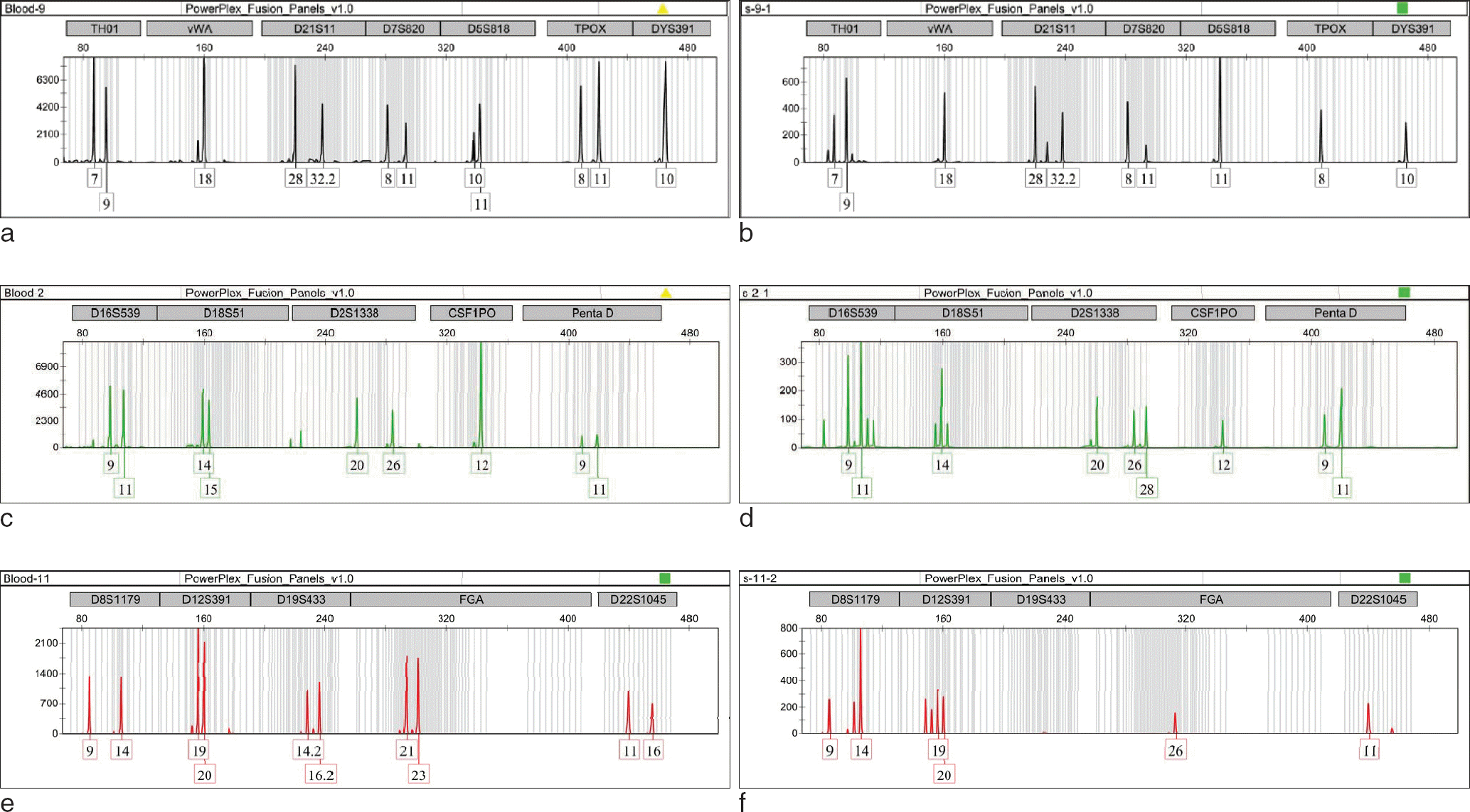 | Fig. 1.The former electropherogram (a, c, e) obtained from blood, and the latter (b, d, f) obtained from serum. (b) indicates allele drop-out of the serum's 11 allele at locus TPOX and 10 allele at locus D5S818. (d) indicates allele drop-in of the serum's 28 allele at locus D2S1338 and allele drop-out of the serum's 15 allele at locus D18S51. (f) indicates drop-in of the serum's 26 allele and drop-out of the 21, 23 allele at locus D22S1045. Also indicates unamplified at locus D19S433. |
Table 1.
The Number of Amplified Allele and the Percentage of Drop-out, Drop-in Ratio




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download