Abstract
To estimate the postmortem interval (PMI) by using entomological evidence, species identification of forensically important flies is mandatory. However, the traditional species identification method, which relies on the key morphological features of adult flies, is not always available to investigators and has limitations to the immature samples. Because of these limitations, species identification using DNA sequences has long been an issue in the field of forensic entomology. In this review, I have briefly described the basic principles of molecular species identification and phylogenetic analysis and their applications in forensic entomology. I also recommend an experimental and statistical method to identify unknown fly samples obtained from the field.
Go to : 
REFERENCES
1. Gennard DE. Forensic entomology: an introduction. 2nd ed.Chichester: John Wiley & Sons;2012.
2. Sperling FA, Anderson GS, Hickey DA. A DNA-based approach to the identification of insect species used for postmortem interval estimation. J Forensic Sci. 1994; 39:418–27.
3. Singh B, Wells JD. Molecular systematics of the Calliphoridae (Diptera: Oestroidea): evidence from one mitochondrial and three nuclear genes. J Med Entomol. 2013; 50:15–23.

4. Zaidi F, Wei Sj, Shi M, et al. Utility of multi-gene loci for forensic species diagnosis of blowflies. J Insect Sci. 2011; 11:1–12.

5. Boehme P, Amendt J, Zehner R. The use of COI barcodes for molecular identification of forensically important fly species in Germany. Parasitol Res. 2012; 110:2325–32.

6. Cognato AI. Standard percent DNA sequence difference for insects does not predict species boundaries. J Econ Entomol. 2006; 99:1037–45.

7. Wells JD, Lunt N, Villet MH. Recent African derivation of Chrysomya putoria from C. chloropyga and mitochondrial DNA paraphyly of cytochrome oxidase subunit one in blowflies of forensic importance. Med Vet Entomol. 2004; 18:445–8.

8. Kumar S, Nei M, Dudley J, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008; 9:299–306.

9. Kumar S, Tamura K, Nei M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci. 1994; 10:189–91.

10. Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007; 24:1596–9.

11. Graur D, Li W-H. Fundamentals of molecular evolution. 2nd ed.Sunderland: Sinauer Associates;2000.
12. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian infer-ence of phylogenetic trees. Bioinformatics. 2001; 17:754–5.

13. Caine ′ LM, Real FC, Salon ̃a-Bordas MI, et al. DNA typing of Diptera collected from human corpses in Portugal. Forensic Sci Int. 2009; 184:e21–3.
14. Chen WY, Hung TH, Shiao SF. Molecular identification of forensically important blow fly species (Diptera: Calliphoridae) in Taiwan. J Med Entomol. 2004; 41:47–57.

15. Debry RW, Timm AE, Dahlem GA, et al. mtDNA-based identification of Lucilia cuprina (Wiedemann) and Lucilia sericata (Meigen) (Diptera: Calliphoridae) in the continental United States. Forensic Sci Int. 2010; 202:102–9.

16. GilArriortua M, Salona Bordas MI, Caine ′ LM, et al. Cytochrome b as a useful tool for the identification of blowflies of forensic interest (Diptera, Calliphoridae). Forensic Sci Int. 2013; 228:132–6.

17. Guo YD, Cai JF, Li X, et al. Identification of the forensically important sarcophagid flies Boerttcherisca peregrina, Parasarcophaga albiceps and Parasarcophaga dux (Diptera: Sarcophagidae) based on COII gene in China. Trop Biomed. 2010; 27:451–60.
18. Harvey ML, Dadour IR, Gaudieri S. Mitochondrial DNA cytochrome oxidase I gene: potential for distinction between immature stages of some forensically important fly species (Diptera) in western Australia. Forensic Sci Int. 2003; 131:134–9.

19. Jordaens K, Sonet G, Richet R, et al. Identification of forensically important Sarcophaga species (Diptera: Sarcophagidae) using the mitochondrial COI gene. Int J Legal Med. 2013; 127:491–504.

20. Kavitha R, Nazni WA, Tan TC, et al. Molecular identification of blow flies recovered from human cadavers during crime scene investigations in Malaysia. Malays J Pathol. 2012; 34:127–32.
21. Meiklejohn KA, Wallman JF, Dowton M. DNA-based identification of forensically important Australian Sarcophagidae (Diptera). Int J Legal Med. 2011; 125:27–32.

22. Meiklejohn KA, Wallman JF, Dowton M. DNA barcoding identifies all immature life stages of a forensically important flesh fly (Diptera: Sarcophagidae). J Forensic Sci. 2013; 58:184–7.

23. Park SH, Zhang Y, Piao H, et al. Use of cytochrome c oxidase subunit i (COI) nucleotide sequences for identification of the Korean Luciliinae fly species (Diptera: Calliphoridae) in forensic investigations. J Korean Med Sci. 2009; 24:1058–63.

24. Park SH, Zhang Y, Piao H, et al. Sequences of the cytochrome C oxidase subunit I (COI) gene are suitable for species identification of Korean Calliphorinae flies of forensic importance (Diptera: Calliphoridae). J Forensic Sci. 2009; 54:1131–4.

25. Saigusa K, Matsumasa M, Yashima Y, et al. Practical applications of molecular biological species identification of forensically important flies. Leg Med (Tokyo). 2009; 11(Suppl 1):S344–7.

26. Saigusa K, Takamiya M, Aoki Y. Species identification of the forensically important flies in Iwate prefecture, Japan based on mitochondrial cytochrome oxidase gene subunit I (COI) sequences. Leg Med (Tokyo). 2005; 7:175–8.

27. Stevens J, Wall R. Species, sub-species and hybrid populations of the blowflies Lucilia cuprina and Lucilia sericata (Diptera: Calliphoridae). Proc Biol Sci. 1996; 263:1335–41.
28. Stevens JR, Wall R, Wells JD. Paraphyly in Hawaiian hybrid blowfly populations and the evolutionary history of anthropophilic species. Insect Mol Biol. 2002; 11:141–8.

29. Wallman JF, Donnellan SC. The utility of mitochondrial D-NA sequences for the identification of forensically important blowflies (Diptera: Calliphoridae) in southeastern Australia. Forensic Sci Int. 2001; 120:60–7.

30. Wells JD, Pape T, Sperling FA. DNA-based identification and molecular systematics of forensically important Sarcophagidae (Diptera). J Forensic Sci. 2001; 46:1098–102.

31. Wells JD, Sperling FA. A DNA-based approach to the identification of insect species used for postmortem interval estimation and partial sequencing of the cytochrome oxy-dase b subunit gene I: a tool for the identification of European species of blow flies for postmortem interval estimation. J Forensic Sci. 2000; 45:1358–9.
32. Wells JD, Wall R, Stevens JR. Phylogenetic analysis of forensically important Lucilia flies based on cytochrome oxidase I sequence: a cautionary tale for forensic species determination. Int J Legal Med. 2007; 121:229–33.

33. Sonet G, Jordaens K, Braet Y, et al. Why is the molecular identification of the forensically important blowfly species Lucilia caesar and L. illustris (family Calliphoridae) so problematic? Forensic Sci Int. 2012; 223:153–9.
34. Piao HG, Chung U, Shin SE, et al. DNA-based identification of necrophagous fly species using abdominal-B (Abd-B) homeobox sequence. Korean J Leg Med. 2012; 36:74–84.
35. Park SH, Park CH, Zhang Y, et al. Using the developmental gene bicoid to identify species of forensically important blowflies (Diptera: Calliphoridae). Biomed Res Int. 2013. DOI: doi: 10.1155/2013/538051. Epub 2013 Mar 18.
36. Zhang Y. Identification of carrion flies by ultrabithorax (Ubx) and abdominal-A (abd-A) homeobox. Seoul: Korea University;2007.
37. Park SH. Molecular identification of necrophagous fly species by sequence analysis of Dfd and Scr homeoboxes. Seoul: Korea University;2007.
Go to : 
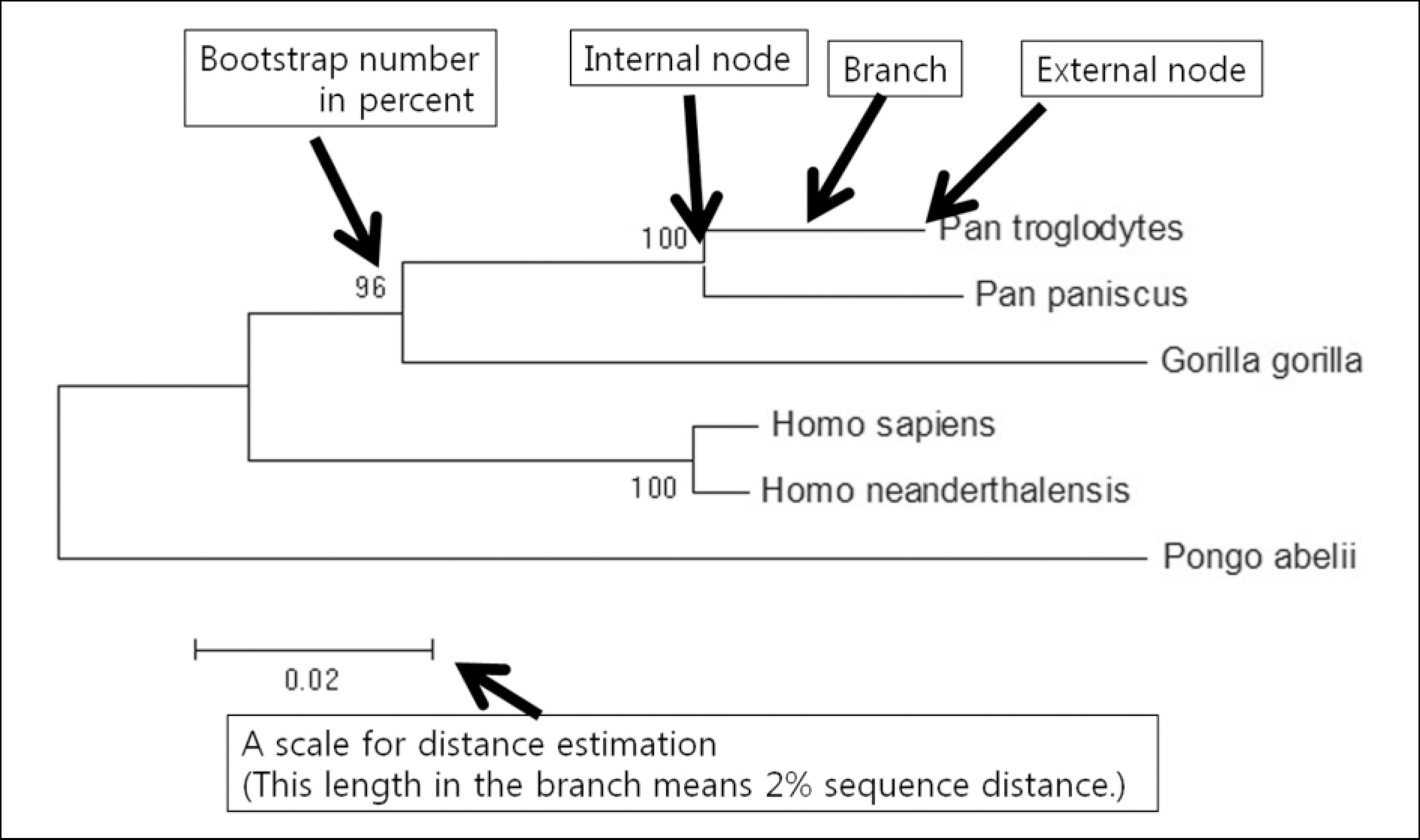 | Fig. 1.An example of a neighbor-joining phylogenetic tree, using COI sequences of modern human (Homo sapiens), Neanderthal man (Homo neanderthalensis), chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (Gorilla gorilla), and orangutan (Pongo abelii), illustrates the key features of a phylogenetic tree such as nodes, branches, bootstrap numbers, and the scale. |
Table 1.
List of Forensically Important Fly Species Commonly Collected in Korea
| Family | Subfamily | Species | Genbank |
|---|---|---|---|
| Fanniidae | Fannia prisca | JX861413-7 | |
| Muscidae | Azelinae | Hydrotaea dentipes | JX861420-JX861429 |
| Hydrotaea occulta | JX861430 | ||
| Ophyra chalcogaster | JX861452-JX861456 | ||
| Ophyra leucostoma | JX861457-JX861459 | ||
| Ophyra nigra | JX861460-JX861468 | ||
| Muscina angustifrons | JX861436-JX861444 | ||
| Muscina stabulans | JX861449-JX861451 | ||
| Muscina pascuorum | JX861445-JX861448 | ||
| Muscinae | Musca domestica | JX861431-JX861435 | |
| Phaoniinae | Phaonia aurea | JX861481&JX861482 | |
| Calliphoridae | Calliphorinae | Adrichina grahami | EU880180-EU880182 |
| Calliphora lata | EU880183-EU880187 | ||
| Calliphora vicina | EU880188-EU880192 | ||
| Triceratopyga calliphoroides | EU880176-EU880179 | ||
| Chrysomyinae∗ | Chrysomya megacephala | KF037969 | |
| Chrysomya pinguis | FJ195381 | ||
| Phormia regina | JN257239&FJ360867 | ||
| Luciliinae | Hemipyrellia ligurriens | EU880206&EU880207 | |
| Lucilia ampullaceae | EU925394 | ||
| Lucilia caesar | EU880193-EU880196 | ||
| Lucilia illustris | EU880197-EU880205 | ||
| Phaenicia sericata | EU880208-EU880212 | ||
| Sarcophagidae | Sarcophaginae | Sarcophaga haemorrhoidalis | JX861406-JX861408 |
| Boettcherisca peregrina | JX861409-JX861412 | ||
| Helicophagella melanura | JX861418&JX861419 | ||
| Parasarcophaga albiceps | JX861469-JX861473 | ||
| Parasarcophaga harpax | JX861474&JX861475 | ||
| Parasarcophaga similis | JX861476-JX861480 |
Table 2.
Universal Primer Sequences for COI∗
| Name | Sequence | Binding site |
|---|---|---|
| F1 5 | 5′ -CCTTTAGAATTGCAGTCTAATGTCA-3′ | tRNA-cysteine |
| F2 5 | 5′ -GGAGGATTTGGAAATTGATTAGTTCC-3′ | ′220-245 on COI |
| F3 5 | 5′ -CTGCTACTTTATGAGCTTTAGG-3′ | 1000-1022 on COI |
| R1 5 | 5′ -CCTAAATTTGCTCATGTTGACA-3′ | 2-23 on COII† |
| R2 5 | 5′ -CAAGTTGTGTAAGCATC-3′ | 1327-1343 on COI |
| R3 5 | 5′ -CCAAAGAATCAAAATAAATGTTG-3′ | 688-710 on COI |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download