Abstract
We developed a web-based Y chromosomal short tandem repeat (Y-STR) database (ySTRmanager, http://ystrmanager.yonsei.ac.kr) to facilitate calculation of Y-STR haplotype frequency estimates for random matches and kinship indices for various relationship levels. The ySTRmanager database provides 3 functions: (i) Y-STR haplotype search, (ii) kinship index calculation, and (iii) user database configuration. The Y-STR haplotype search function allows researchers to retrieve Y-STR haplotypes that meet queried Y-STR allele, Y-haplogroup affiliation, and/or sample information from a selected population in the open database, which consists of 12-17 Y-STR loci. The number of matches in a selected population, haplotype frequency estimator, and detailed results for matched and neighbor haplotypes are displayed as a set of search results. The kinship index calculation function provides kinship indices of 2 input Y-STR haplotypes for the relationship represented by the number of meioses, with consideration of target population and mutation rate of each Y-STR. In addition, ySTRmanager allows registered users to configure their own database to store and analyze Y-STR haplotype and/or mutation rate data. The stored Y-STR data can be used in the search function and in the analysis to obtain forensic statistical values. The ySTRmanager will be a useful system to analyze and manage Y-STR data in the practice of forensic genetics.
Figures and Tables
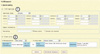 | Fig. 1Search setting in ySTRmanager. In order to search a particular Y-STR haplotype (Y-haplotype) from a selected population, user should provide Y-haplotype information (circled "1") and then select a target population (circled "2"). Y-STR allele values can be provided by selecting a value in list box for each Y-STR loci interested or by direct input of allele values into text box after selecting "Others" in list box. Y-haplogroup and/or sample information can be combined to Y-haplotype or used independently to search haplotypes in a selected population. |
 | Fig. 2Search results of ySTRmanager. The number of matches for the particular haplotype and its haplotype frequency estimator in a selected population are displayed in search result (the top table of a). Individual detailed information including sample name, Y-haplogroup affiliation, and sample information for matched haplotypes are shown in a table, which can be downloaded by clicking "Export" button (the bottom table of a). Neighbor haplotypes are displayed in a table only when there is at least one matched haplotype in a selected population (b). Allele values at Y-STR loci with single-step mutations (+1 repeat gain or -1 repeat loss) in neighbor haplotypes are represented by different backgrounds (dashed box in b). Detailed sample information having the neighbor haplotypes can also be appeared in a new window by clicking "View" link. |
References
1. Roewer L, Arnemann J, Spurr NK, Grzeschik KH, Epplen JT. Simple repeat sequences on the human Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet. 1992. 89:389–394.
2. Jobling MA, Pandya A, Tyler-Smith C. The Y chromosome in forensic analysis and paternity testing. Int J Legal Med. 1997. 110:118–124.
3. Kayser M, Caglià A, Corach D, et al. Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med. 1997. 110:125–133. appendix 141-9.
4. de Knijff P, Kayser M, Caglià A, et al. Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Legal Med. 1997. 110:134–149.
5. Jobling MA, Tyler-Smith C. Fathers and sons: the Y chromosome and human evolution. Trends Genet. 1995. 11:449–456.
6. Roewer L, Kayser M, de Knijff P, et al. A new method for the evaluation of matches in non-recombining genomes: application to Y-chromosomal short tandem repeat (STR) haplotypes in European males. Forensic Sci Int. 2000. 114:31–43.
7. Willuweit S, Roewer L. International Forensic Y Chromosome User Group. Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007. 1:83–87.
8. Ge J, Budowle B, Planz JV, Eisenberg AJ, Ballantyne J, Chakraborty R. US forensic Y-chromosome short tandem repeats database. Leg Med (Tokyo). 2010. 12:289–295.
9. Rolf B, Keil W, Brinkmann B, Roewer L, Fimmers R. Paternity testing using Y-STR haplotypes: assigning a probability for paternity in cases of mutations. Int J Legal Med. 2001. 115:12–15.
10. Kurihara R, Yamamoto T, Uchihi R, et al. Mutations in 14 Y-STR loci among Japanese father-son haplotypes. Int J Legal Med. 2004. 118:125–131.
11. Lee HY, Park MJ, Chung U, et al. Haplotypes and mutation analysis of 22 Y-chromosomal STRs in Korean fatherson pairs. Int J Legal Med. 2007. 121:128–135.
12. Decker AE, Kline MC, Redman JW, Reid TM, Butler JM. Analysis of mutations in father-son pairs with 17 Y-STR loci. Forensic Sci Int Genet. 2008. 2:e31–e35.
13. Goedbloed M, Vermeulen M, Fang RN, et al. Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR Yfiler PCR amplification kit. Int J Legal Med. 2009. 123:471–482.
14. Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934. 26:404–413.
15. Buckleton JS, Krawczak M, Weir BS. The interpretation of lineage markers in forensic DNA testing. Forensic Sci Int Genet. 2011. 5:78–83.
16. Buckleton JS, Triggs CM, Walsh SJ. Forensic DNA evidence interpretation. 2005. 1st ed. Boca Raton: CRC press;388–389.
17. Summary list of Y chromosome STR loci and available fact sheets. Available from: http://www.cstl.nist.gov/strbase/ystr_fact.htm.
18. Park MJ, Lee HY, Kim NY, et al. Genetic characteristics of 22 Y-STR loci in Koreans. Korean J Leg Med. 2007. 31:162–170.
19. Kim SH, Kim NY, Hong SB, et al. Genetic polymorphisms of 16 Y chromosomal STR loci in Korean population. Forensic Sci Int Genet. 2008. 2:e9–e10.
20. Seong KM, Yoo SY, Hwang JH, Kim SH, Chung KW, Cho NS. Population genetic polymorphisms of 17 Y-chromosomal STR loci in South Koreans. Forensic Sci Int Genet. 2011. 5:e122–e123.
21. Hu SP. Polymorphism of Y-chromosomal STR haplotypes in the Chaoshan Han Chinese in South China. Forensic Sci Int. 2006. 158:80–85.
22. Hu SP. Genetic polymorphism of 12 Y-chromosomal STR loci in the Minnan Han Chinese in Southeast China. Forensic Sci Int. 2006. 159:77–82.
23. Zhu B, Deng Y, Zhang F, et al. Genetic analysis for Y chromosome short tandem repeat haplotypes of Chinese Han population residing in the Ningxia province of China. J Forensic Sci. 2006. 51:1417–1420.
24. Yang B, Gu M, Wang G, Li X, Liu Y, Yang W. Population data for 11 Y-chromosome STRs in northeast China Han. Forensic Sci Int. 2006. 164:65–71.
25. Yan J, Tang H, Liu Y, et al. Genetic polymorphisms of 17 Y-STRs haplotypes in Chinese Han population residing in Shandong province of China. Leg Med (Tokyo). 2007. 9:196–202.
26. Zhang H, Yun L, Li Y, et al. Haplotype of 12 Y-STR loci of the PowerPlex Y-system in Sichuan Han ethnic group in west China. Forensic Sci Int. 2008. 175:244–249.
27. Yanmei Y, Tao G, Yubao Z, et al. Genetic polymorphism of 11 Y-chromosomal STR loci in Yunnan Han Chinese. Forensic Sci Int Genet. 2010. 4:e67–e69.
28. Guo H, Yan J, Jiao Z, et al. Genetic polymorphisms for 17 Y-chromosomal STRs haplotypes in Chinese Hui population. Leg Med (Tokyo). 2008. 10:163–169.
29. Zhang Y, Zhang H, Cui Y, et al. Population genetics for Y-chromosomal STRs haplotypes of Chinese Korean ethnic group in northeastern China. Forensic Sci Int. 2007. 173:197–203.
30. Bai R, Shi M, Yu X, Zhang J, Bai L. Y-chromosomal STRs haplotypes in Chinese Manchu ethnic group. Forensic Sci Int Genet. 2008. 3:e13–e15.
31. Xin N, Chen T, Yu B, Li S. 12 Y-STRs haplotypes in Chinese Naxi ethnic minority group. Forensic Sci Int. 2008. 174:244–248.
32. Zhu B, Shen C, Xun X, et al. Population genetic polymorphisms for 17 Y-chromosomal STRs haplotypes of Chinese Salar ethnic minority group. Leg Med (Tokyo). 2007. 9:203–209.
33. Yi Z, Jun W, XingBo S, XiaoJun L, Liu D, BinWu Y. Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic minority group. Leg Med (Tokyo). 2010. 12:108–111.
34. Shi M, Bai R, Wan L, Yu X, Chang L. Population genetics for Y-chromosomal STRs haplotypes of Chinese Tujia ethnic group. Forensic Sci Int Genet. 2008. 2:e65–e68.
35. Zhu B, Shen C, Qian G, et al. Genetic polymorphisms for 11 Y-STRs haplotypes of Chinese Yi ethnic minority group. Forensic Sci Int. 2006. 158:229–233.
36. Mizuno N, Nakahara H, Sekiguchi K, Yoshida K, Nakano M, Kasai K. 16 Y chromosomal STR haplotypes in Japanese. Forensic Sci Int. 2008. 174:71–76.
37. Hashiyada M, Umetsu K, Yuasa I, et al. Population genetics of 17 Y-chromosomal STR loci in Japanese. Forensic Sci Int Genet. 2008. 2:e69–e70.
38. Hwa HL, Tseng LH, Ko TM, et al. Seventeen Y-chromosomal short tandem repeat haplotypes in seven groups of population living in Taiwan. Int J Legal Med. 2010. 124:295–300.
39. Wu FC, Ho CW, Pu CE, et al. Y-chromosomal STRs haplotypes in the Taiwanese Paiwan population. Int J Legal Med. 2011. 125:39–43.
40. Chang YM, Perumal R, Keat PY, Kuehn DL. Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forensic Sci Int. 2007. 167:70–76.
41. Yong RY, Lee LK, Yap EP. Y-chromosome STR haplotype diversity in three ethnic populations in Singapore. Forensic Sci Int. 2006. 159:244–257.
42. Berger B, Lindinger A, Niederstätter H, Grubwieser P, Parson W. Y-STR typing of an Austrian population sample using a 17-loci multiplex PCR assay. Int J Legal Med. 2005. 119:241–246.
43. Hallenberg C, Nielsen K, Simonsen B, Sanchez J, Morling N. Y-chromosome STR haplotypes in Danes. Forensic Sci Int. 2005. 155:205–210.
44. Rodig H, Roewer L, Gross A, et al. Evaluation of haplotype discrimination capacity of 35 Y-chromosomal short tandem repeat loci. Forensic Sci Int. 2008. 174:182–188.
45. Völgyi A, Zalán A, Szvetnik E, Pamjav H. Hungarian population data for 11 Y-STR and 49 Y-SNP markers. Forensic Sci Int Genet. 2009. 3:e27–e28.
46. Turrina S, Atzei R, De Leo D. Y-chromosomal STR haplotypes in a Northeast Italian population sample using 17plex loci PCR assay. Int J Legal Med. 2006. 120:56–59.
47. Soltyszewski I, Pepinski W, Spolnicka M, Kartasinska E, Konarzewska M, Janica J. Y-chromosomal haplotypes for the AmpFlSTR Yfiler PCR Amplification Kit in a population sample from Central Poland. Forensic Sci Int. 2007. 168:61–67.
48. Alves C, Gomes V, Prata MJ, Amorim A, Gusmão L. Population data for Y-chromosome haplotypes defined by 17 STRs (AmpFlSTR YFiler) in Portugal. Forensic Sci Int. 2007. 171:250–255.
49. Pontes ML, Cainé L, Abrantes D, Lima G, Pinheiro MF. Allele frequencies and population data for 17 Y-STR loci (AmpFlSTR Y-filer) in a Northern Portuguese population sample. Forensic Sci Int. 2007. 170:62–67.
50. Valverde L, Köhnemann S, Rosique M, et al. 17 Y-STR haplotype data for a population sample of Residents in the Basque Country. Forensic Sci Int Genet. 2012. In press.
51. Roewer L, Willuweit S, Krüger C, et al. Analysis of Y chromosome STR haplotypes in the European part of Russia reveals high diversities but non-significant genetic distances between populations. Int J Legal Med. 2008. 122:219–223.
52. Veselinovic IS, Zgonjanin DM, Maletin MP, et al. Allele frequencies and population data for 17 Y-chromosome STR loci in a Serbian population sample from Vojvodina province. Forensic Sci Int. 2008. 176:e23–e28.
53. Martín P, García-Hirschfeld J, García O, et al. A Spanish population study of 17 Y-chromosome STR loci. Forensic Sci Int. 2004. 139:231–235.
54. Sánchez C, Barrot C, Xifró A, et al. Haplotype frequencies of 16 Y-chromosome STR loci in the Barcelona metropolitan area population using Y-Filer kit. Forensic Sci Int. 2007. 172:211–217.
55. Haas C, Wangensteen T, Giezendanner N, Kratzer A, Bär W. Y-chromosome STR haplotypes in a population sample from Switzerland (Zurich area). Forensic Sci Int. 2006. 158:213–218.
56. Ballard DJ, Phillips C, Thacker CR, Robson C, Revoir AP, Syndercombe Court D. Y chromosome STR haplotypes in three UK populations. Forensic Sci Int. 2005. 152:289–305.
57. Decker AE, Kline MC, Redman JW, Reid TM, Butler JM. Analysis of mutations in father-son pairs with 17 Y-STR loci. Forensic Sci Int Genet. 2008. 2:e31–e35.
58. Marino M, Furfuro S. Genetic population data of 12 Y-chromosome STRs loci in Mendoza population (Argentina). Forensic Sci Int Genet. 2010. 4:e89–e93.
59. Pereira RW, Monteiro EH, Hirschfeld GC, Wang AY, Grattapaglia D. Haplotype diversity of 17 Y-chromosome STRs in Brazilians. Forensic Sci Int. 2007. 171:226–236.
60. Builes JJ, Bravo ML, Gómez C, et al. Y-chromosome STRs in an Antioquian (Colombia) population sample. Forensic Sci Int. 2006. 164:79–86.
61. Builes JJ, Martínez B, Gómez A, et al. Y chromosome STR haplotypes in the Caribbean city of Cartagena (Colombia). Forensic Sci Int. 2007. 167:62–69.
62. Baeza C, Guzmán R, Tirado M, et al. Population data for 15 Y-chromosome STRs in a population sample from Quito (Ecuador). Forensic Sci Int. 2007. 173:214–219.
63. Matamoros M, Yurrebaso I, Gusmão L, García O. Population data for 12 Y-chromosome STR loci in a sample from Honduras. Leg Med (Tokyo). 2009. 11:251–255.
64. Luna-Vázquez A, Vilchis-Dorantes G, Aguilar-Ruiz MO, et al. Haplotype frequencies of the PowerPlex Y system in a Mexican-Mestizo population sample from Mexico City. Forensic Sci Int Genet. 2008. 2:e11–e13.
65. Borjas L, Bernal LP, Chiurillo MA, et al. Usefulness of 12 Y-STRs for forensic genetics evaluation in two populations from Venezuela. Leg Med (Tokyo). 2008. 10:107–112.
66. Willuweit S, Caliebe A, Andersen MM, Roewer L. Y-STR Frequency Surveying Method: a critical reappraisal. Forensic Sci Int Genet. 2011. 5:84–90.
67. Holland MM, Parsons TJ. Mitochondrial DNA sequence analysis - validation and use for forensic case work. Forensic Sci Rev. 1999. 11:21–50.
68. Ota T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973. 22(2):201–204.
69. Fan H, Chu JY. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinformatics. 2007. 5(1):7–14.
70. Vermeulen M, Wollstein A, van der Gaag K, et al. Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forensic Sci Int Genet. 2009. 3(4):205–213.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download