Abstract
The metabolic syndrome refers to a well defined group of risk factors, including central obesity and inflammation, for the development of diabetes and cardiovascular disease. Interestingly, many studies have recently led to the emergence of somewhat unexpected relationships between several infectious diseases and various aspects of the metabolic syndrome. Our understanding of the mechanisms underlying these interactions is also rapidly developing and some of these are summarized in this article. We will focus first on bacterial infection, and most notably the role of gut microbiota in regulaton of both obesity and inflammation. In particular, we focus on the role of inflammasomes and propose that understanding the role of Toll-like receptors and Nod-like receptors in the pathogenesis of inflammatory disorders with or without infection may provide novel targets for prevention and/or treatment of associated diseases. Secondly, chronic bacterial or viral infection and emerging links with metabolism will be reviewed. Finally, consideratons of biomarkers for metabolic syndrome, in particular lipocalin-2, and their link with infection will be discussed.
The metabolic syndrome describes a cluster of risk factors for the development of diabetes and cardiovascular disease. The latest clinical definition of the metabolic syndrome by the International Diabetes Federation and the American Heart Association/National Heart, Lung, and Blood Institute [1] states that three abnormal findings in the following five would qualify a person as having the metabolic syndrome - raised blood pressure, atherogenic dyslipidemia, raised triglycerides, low high density lipoprotein cholesterol (HDL-C), dysglycemia, and central obesity (excessive fat tissue in and around the abdomen). Of course the prognosis afforded by these considerations is not absolute since additional factors such as age, gender and lifestyle (e.g., smoking, level of exercise) contribute to absolute risk. The prevalence of the metabolic syndrome has increased steadily worldwide and in the United States an estimated 50 million Americans fall in this category. Associated with this is clearly a profound impact on morbidity and mortality.
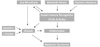
Obesity and the metabolic syndrome are associated with a proinflammatory state [2], often defined by elevated circulating C-reactive protein. Studies on the central role of inflammation have recently unearthed many rather unexpected nodes of interaction with various infectious diseases, previously thought to be unrelated to metabolic syndrome (Fig. 1). For example, there are now clear interactions in particular between gut microbiota as well as chronic bacterial or viral infection and various aspects of the metabolic syndrome such as obesity and inflammation. Our understanding of the mechanisms underlying these interactions is also rapidly developing and recent data and potential future studies are described in more detail below.
The human gut is populated by at least 1013 microorganisms, mostly anaerobic bacteria, generally known as the microbiota. The metabolic activities of these microbes are comparable to an 'organ' particularly adapted to human physiology and executing vital functions including the ability to process otherwise indigestible nutrients, repressing the growth of harmful microorganisms and training the immune system to respond only to pathogens [3]. In recent years it became evident that the gut microbiota constitutes an important node for integrating environmental factors and metabolic processes in the host, including energy extraction from nutrients and fat storage. This suggested the relatively new concept of a possible role in obesity and, consequently, other aspects of the metabolic syndrome [4]. The two predominant populations of microbiota in both rodent and human gut are members of the bacterial groups known as the Firmicutes and the Bacteroidetes and the relative proportion of these two phyla may protect or predispose the host to obesity [5]. Metagenomic studies demonstrated that the proportion of Firmicutes is higher in obese individuals as compared to lean controls and this correlates with a higher number of genes encoding enzymes that break down otherwise indigestible dietary polysaccharides, more fermentation end products and fewer calories remaining in the faeces of obese individuals. Intriguingly, microbiota transplantation studies in germ-free murine models showed that the efficient energy extraction traits of obese-type gut flora are transmissible [6,7]. Although such experiments are an extreme example they clearly show that obesity has 'infectious' characteristics. Collectively these observations indicate that differences in the efficiency of caloric extraction from food may be determined by the composition of the microbiota, which, in turn, may contribute to differential body weights. Another mechanism by which the microbiome may contribute to metabolic disorders is via triggering systemic inflammation, as described below.
The gastrointestinal tract of a normal fetus is sterile. During birth and rapidly thereafter, bacteria from the mother and the surrounding environment colonize the infant's gut. As a consequence, the immune system coevolves with the microbiota during postnatal life, which allows the host and microbiota to coexist in a mutually beneficial relationship. In particular the innate immune system, long-appreciated for its role in defending against infection by pathogenic microbes, has emerged as a key regulator of the gut microbiota. Innate immune recognition of microbe-associated molecular patterns by multiple families of pattern-recognition molecules such as Toll-like receptors (TLRs) and Nod-like receptors (NLRs) instructs the innate and adaptive immune system to protect the host from pathogens while also acting to establish a beneficial mutualism with commensal organisms[8,9]. Therefore, molecular integration of nutrient- and pathogen sensing pathways became of great interest in understanding the mechanisms of insulin resistance, type 2 diabetes (T2D), and other chronic metabolic pathologies.
Recent findings indicate that TLRs, which are up-regulated in the affected tissue of most inflammatory disorders, can mediate crosstalk between the immune systems and whole body metabolism [10]. TLR's are a major form of cell surface innate immune sensor and their activation induces the recruitment of cells such as macrophages and neutrophils [11]. It has been demonstrated that TLR4, a sensor for lipopolysaccharides on Gram-negative bacteria, is involved in the induction of proinflammatory cytokine expression in macrophages, adipocytes and liver [12]. TLR4 is also activated in response to free fatty acids, which are known to play a key role in the etiology of insulin resistance. Importantly, mice genetically deficient for TLR4 are substantially protected from the ability of systemic lipid infusion to suppress insulin signaling in muscle and reduce insulin-mediated changes in systemic glucose metabolism [12]. Similarly, it was recently reported that mice genetically deficient in TLR5, which recognizes flagellin (the protein monomer that makes up the filament of bacterial flagella, found on nearly all motile bacteria) exhibit hyperphagia and develop hallmark features of metabolic syndrome, including hyperlipidemia, hypertension, insulin resistance, and increased adiposity [13]. While the underlying molecular mechanisms remain to be defined, it is tempting to speculate that loss of TLR5 produces alterations in the gut microbiota that induce low-grade inflammatory signaling. This signaling may in turn decrease insulin sensitivity, induce hyperphagia, and ultimately initiate development of various other aspects of metabolic syndrome.
NLR signaling results in the formation of large molecular scaffold complexes with multiple components (termed an inflammasome) which are intricately linked with inflammation and crosstalk with TLR-mediated signaling events [11]. NLR family inflammasomes have emerged as central platforms of innate immunity that link the sensing of infectious agents and metabolic stress to the activation of pro-inflammatory cytokines such as interleukin (IL)-1β and IL-18. Inflammasomes do not regulate transcription of immune response genes but instead activate caspase-1, an integral component of the inflammasome and proteolytic enzyme that cleaves and activates these secreted cytokines [14]. Currently the most fully characterized inflammasome, the NLRP3 inflammasome consists of the NLRP3 scaffold, the ASC (PYCARD) adaptor, and caspase-1, which is activated upon a number of diverse stimuli and dysregulated in various diseases [15]. Thus, coordinated inputs from TLR (gene transcription) and NLR (processing) are often needed for full activation of one of the most important proinflammatory cytokines, IL-1β [16].
Additional functions of NLR inflammasomes continue to emerge. Inflammasomes regulate pyroptosis, a caspase-1-dependent form of cell death that itself is highly inflammatory [17]. Intriguingly, the NLRP3 inflammasome also detects signs of metabolic stress, including elevated extracellular glucose [18], Thus, it has been demonstrated that NLRP3 interacts with thioredoxin (TRX)-interacting protein (TXNIP), a protein linked to insulin resistance. TXNIP deficiency impaired activation of the NLRP3 inflammasome and subsequent activation of IL-1β [15]. It is established that IL-1β plays an important role in mediating the toxic effects of prolonged hyperglycaemia (glucotoxicity) in pancreatic islets, driving β cell destruction and dysregulating glucose-stimulated insulin secretion [19]. Thus, it is important to note that mice genetically deficient for both TXNIP and NLRP3 showed impaired glucose homeostasis, providing further evidence for a mechanistic link between IL-1β in the pathogenesis of diabetes [15]. The therapeutic potential of targeting the mechanisms outlined above are reinforced by the recent promise of IL-1β receptor antagonism in clinical trials for the treatment of diabetes [20,21].
As activation of inflammasomes emerges as a central event in sensing exogenous and endogenous danger signals, it appears also worthwhile to reconsider aspects of metabolic dysfunction with concurrent chronic infection. It is also conceivable that chronic bacterial and viral infections trigger pro-inflammatory signals, which may induce metabolic pathologies. For example, there is evidence that individuals infected with multiple pathogens such as HSV-1, HSV-2, CMV, Helicobacter pylori, and hepatitis A have high C-reactive protein levels, indicating enhanced inflammation [22]. These individuals also had an elevated relative risk for coronary artery disease. A cross-sectional population-based study reported a significant positive correlation of various parameters defining the metabolic syndrome and chronic infection with Chlamydia pneumoniae, Helicobacter pylori, CMV and HSV-1 [23]. Moreover, human immunodeficiency virus (HIV)-infected patients develop multiple metabolic abnormalities including insulin resistance, lipodystrophy and dyslipidemia. Although this is often attributed to therapy, it is now clear that lipid and glucose abnormalities are apparent in HIV-infected patients even before commencing highly active antiretroviral therapy (HAART) [24]. Another striking example is the hepatitis C virus (HCV), which induces inflammation via several mechanisms and ultimately leads to insulin resistance, steatosis, fibrosis, apoptosis and hepatocellular carcinoma. Proteomic data suggest that HCV induces early perturbations in glycolysis, the pentose phosphate pathway, and the citric acid cycle, which favor host biosynthetic activities supporting viral replication and propagation. This is followed by a compensatory shift in metabolism aimed at maintaining energy homeostasis and cell viability during elevated viral replication and increasing cellular stress [25]. With emerging insight into the pathogenic mechanisms leading to insulin resistance in infected individuals, HCV is now regarded as a cause of metabolic syndrome-like outcomes, as opposed to simple viral infection [26]. In a similar fashion, recent evidence demonstrated that HIV replication in human T-cells, without any influence of antiviral drugs or other factors, can stimulate production of novel cellular enzymes and proteins that enhance fatty acid synthesis, increase quantity of low density lipoproteins, secrete triglycerides, dysregulate lipid transport, oxidize lipids, and alter lipid metabolism and therefore may directly impact on lipid synthesis, transport and metabolism [27]. Thus, significant attention is presently being drawn toward the integration of nutrient- and pathogen-sensing pathways and this should provide fertile ground for future mechanistic research and translation of such studies to novel urgently needed therapeutic strategies.
How can the information described above be integrated with currently used or proposed markers for metabolic syndrome? Many biomarkers have potential merits such as high sensitivity C-reactive protein as an overall marker of inflammation, IL-1β as a final mediator of many detrimental effects on various tissues, and the oligomeric forms of adiponectin as an indicator of metabolic, cardiovascular and perhaps even infectious status [26,28-30]. Another particularly interesting example is lipocalin-2, a proinflammatory adipokine which is typically up-regulated in obese subjects and causally involved in the development of obesity-associated insulin resistance and metabolic abnormalities [31]. It now also appears that there are several emerging links between lipocalin-2 and infectious disease. Patients infected with HIV have been shown to have lower circulating lipocalin-2 levels which were restored by HAART [32]. Lipocalin-2 knockout mice are highly susceptible to bacerial infection [33]. This is associated with the fact that lipocalin-2 binds bacterial enterobactin siderophores to limit bacterial iron acquisition. Another critical component of lipocalin-2 action appears to be related with its endocytosis and targeting, along with binding partners including bacteria, to lysosomes for degradation [34]. It is known that as part of the acute phase response to infection, TLR activation induces production of lipocalin-2 and that treatment of adipocytes or macrophages with LPS, IL-1β or additional inflammatory stimuli which activate NFκB results in a dramatic enhancement of lipocalin-2 production [35,36]. We suggest that studies on the use of lipocalin-2 as a biomarker for metabolic disease are promising but must be cogniscant of the coexistence of any infection, acute or chronic, in each patient.
Figures and Tables
ACKNOWLEDGEMENT
GS and PS acknowledge funding from Ministry of Education Science and Technology (MEST) and GS is further supported by a grant from Astra Zeneca. GS also thanks Canadian Diabetes Association, Heart and Stroke Foundation of Canada and Canadian Institutes of Health Research for funding.
References
1. Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009. 120:1640–1645.
2. Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006. 444:860–867.
3. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977. 31:107–133.
4. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004. 101:15718–15723.
5. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005. 102:11070–11075.
6. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009. 457:480–484.
7. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006. 444:1027–1031.
8. Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006. 7:1250–1257.
9. Fritz JH, Le Bourhis L, Magalhaes JG, Philpott DJ. Innate immune recognition at the epithelial barrier drives adaptive immunity: APCs take the back seat. Trends Immunol. 2008. 29:41–49.
10. Kanczkowski W, Ziegler CG, Zacharowski K, Bornstein SR. Toll-like receptors in endocrine disease and diabetes. Neuroimmunomodulation. 2008. 15:54–60.
11. Fukata M, Vamadevan AS, Abreu MT. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Semin Immunol. 2009. 21:242–253.
12. Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006. 116:3015–3025.
13. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010. 328:228–231.
14. Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009. 19:455–464.
15. Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010. 327:296–300.
16. Netea MG, van de Veerdonk FL, Kullberg BJ, Van der Meer JW, Joosten LA. The role of NLRs and TLRs in the activation of the inflammasome. Expert Opin Biol Ther. 2008. 8:1867–1872.
17. Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009. 27:229–265.
18. Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010. 11:136–140.
19. Maedler K, Dharmadhikari G, Schumann DM, Storling J. Interleukin-1 beta targeted therapy for type 2 diabetes. Expert Opin Biol Ther. 2009. 9:1177–1188.
20. Larsen CM, Faulenbach M, Vaag A, Ehses JA, Donath MY, Mandrup-Poulsen T. Sustained effects of interleukin-1 receptor antagonist treatment in type 2 diabetes. Diabetes Care. 2009. 32:1663–1668.
21. Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007. 356:1517–1526.
22. Zhu J, Quyyumi AA, Norman JE, Csako G, Waclawiw MA, Shearer GM, Epstein SE. Effects of total pathogen burden on coronary artery disease risk and C-reactive protein levels. Am J Cardiol. 2000. 85:140–146.
23. Nabipour I, Vahdat K, Jafari SM, Pazoki R, Sanjdideh Z. The association of metabolic syndrome and Chlamydia pneumoniae, Helicobacter pylori, cytomegalovirus, and herpes simplex virus type 1: the Persian Gulf Healthy Heart Study. Cardiovasc Diabetol. 2006. 5:25.
24. El-Sadr WM, Mullin CM, Carr A, Gibert C, Rappoport C, Visnegarwala F, Grunfeld C, Raghavan SS. Effects of HIV disease on lipid, glucose and insulin levels: results from a large antiretroviral-naive cohort. HIV Med. 2005. 6:114–121.
25. Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010. 6:e1000719.
26. Sheikh MY, Choi J, Qadri I, Friedman JE, Sanyal AJ. Hepatitis C virus infection: molecular pathways to metabolic syndrome. Hepatology. 2008. 47:2127–2133.
27. Rasheed S, Yan JS, Lau A, Chan AS. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS One. 2008. 3:e3003.
28. Alexandraki K, Piperi C, Kalofoutis C, Singh J, Alaveras A, Kalofoutis A. Inflammatory process in type 2 diabetes: the role of cytokines. Ann N Y Acad Sci. 2006. 1084:89–117.
29. Bezante GP, Briatore L, Rollando D, Maggi D, Setti M, Ghio M, Agosti S, Murdaca G, Balbi M, Barsotti A, Cordera R. Hypoadiponectinemia in lipodystrophic HIV individuals: a metabolic marker of subclinical cardiac damage. Nutr Metab Cardiovasc Dis. 2009. 19:277–282.
30. Nystrom T. C-reactive protein: a marker or a player? Clin Sci (Lond). 2007. 113:79–81.
31. Alpizar-Alpizar W, Laerum OD, Illemann M, Ramirez JA, Arias A, Malespin-Bendana W, Ramirez V, Lund LR, Borregaard N, Nielsen BS. Neutrophil gelatinase-associated lipocalin (NGAL/Lcn2) is upregulated in gastric mucosa infected with Helicobacter pylori. Virchows Arch. 2009. 455:225–233.
32. Landro L, Damas JK, Flo TH, Heggelund L, Ueland T, Tjonnfjord GE, Espevik T, Aukrust P, Froland SS. Decreased serum lipocalin-2 levels in human immunodeficiency virus-infected patients: increase during highly active anti-retroviral therapy. Clin Exp Immunol. 2008. 152:57–63.
33. Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006. 103:1834–1839.
34. Halaas O, Steigedal M, Haug M, Awuh JA, Ryan L, Brech A, Sato S, Husebye H, Cangelosi GA, Akira S, Strong RK, Espevik T, Flo TH. Intracellular Mycobacterium avium intersect transferrin in the Rab11(+) recycling endocytic pathway and avoid lipocalin 2 trafficking to the lysosomal pathway. J Infect Dis. 2010. 201:783–792.
35. Cowland JB, Muta T, Borregaard N. IL-1beta-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by IkappaB-zeta. J Immunol. 2006. 176:5559–5566.
36. Shen F, Ruddy MJ, Plamondon P, Gaffen SL. Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol. 2005. 77:388–399.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download