Abstract
We report the first Korean case of lung diseases caused by Mycobacterium abscessus subsp. bolletii in a previously healthy male, except for a previous history of pulmonary tuberculosis and bronchiectasis. All serial isolates are identified as M. abscessus subsp. bolletii by multi-locus sequence analysis based on the hsp65, rpoB, and 16S rRNA fragments. At the genetic level, the isolate has the erm(41) gene with a T28 sequevar, associated with clarithromycin resistance, and no rrl mutation. The isolate is resistant to clarithromycin. Although the symptoms and radiographic findings have improved after combination of antibiotics, the follow-up sputum cultures are persistently positive.
Rapidly growing mycobacteria (RGM) are relatively common pathogens of lung disease caused by nontuberculous mycobacteria (NTM) in Korea1. Of the RGM species, Mycobacterium abscessus complex is most commonly associated with NTM lung disease. Although subspeciation of M. abscessus complex is still controversial, M. abscessus complex is currently divided into three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense and M. abscessus subsp. bolletii2.
In Korea, M. abscessus subsp. bolletii is identified in only 1.3-1.6% of clinical strains of M. abscessus complex3. Although a case with disseminated infection of M. abscessus subsp. bolletii was reported4, there has been no report of NTM lung disease caused by M. abscessus subsp. bolletii in Korea. Here, we describe a case of chronic pulmonary disease caused by M. abscessus subsp. bolletii.
A 66-year-old male was referred to our hospital for management of probable NTM lung disease. He was an ex-smoker and had a history of previous treatment for pulmonary tuberculosis 10 years prior. Six months before his visit to our hospital, the patient reported chronic cough and sputum. He was initially diagnosed with acid-fast bacilli (AFB) smear-positive pulmonary tuberculosis and received isoniazid, rifampin, ethambutol, and pyrazinamide for 6 months. However, AFB smears were persistently positive, and NTM were cultured from his sputum specimens. He was then referred to our hospital.
Physical examination showed that the patient was 172.3 cm tall and weighed 56.1 kg. Laboratory tests results were unremarkable. A human immunodeficiency virus antibody test was negative. The chest radiograph and computed tomography (CT) scan revealed multifocal bronchiectasis and bronchiolitis in both lungs that suggested the nodular bronchiectatic form of NTM lung disease (Figure 1).
The patient's sputum was positive for AFB staining, and NTM were isolated more than three times in a liquid culture system (Bactec MGIT 960 system; BD Diagnostics, Sparks, MD, USA). To identify the etiological agent, bacteria were initially propagated in 7H9 broth (Difco Laboratories, Detroit, MI, USA) supplemented with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (BD Diagnostics) for 7 days at 37℃, sub-cultured in egg-based 3% Ogawa solid media (Shinyang, Seoul, Korea), and genomic DNA was extracted from the cultured bacteria. Initial species identification using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method based on the rpoB gene (Myco-ID; M&D Inc., Wonju, Korea) was M. abscessus complex5.
To speciate the isolates, sequencing analysis of the nearly complete 16S rRNA gene, partial hsp65, and rpoB sequences were performed using GenBank (http://blast.ncbi.nlm.nih.gov/) with the BLAST algorithm6. The 16S rRNA sequences showed 100% identity with M. abscessus subsp. abscessus (GenBank accession no. NR074427), M. abscessus subsp. massiliense (GenBank accession no. NR074421), M. chelonae (GenBank accession no. AY457082), and M. abscessus subsp. bolletii (GenBank accession no. NR043236). The hsp65 and rpoB sequences were 100% identical to those of the M. abscessus subsp. bolletii type strain (GenBank accession nos. FJ607778 and AY859692). Phylogenetic analysis based on the rpoB sequences from the isolate SMC-bol-002 and from those of closely related species within the RGM showed that this strain belonged to M. abscessus subsp. bolletii (Figure 2).
Drug susceptibility testing was performed using a broth microdilution method (Table 1)7. At the genetic level, the isolate was genotyped for erm(41) polymorphism and for rrl mutations; these are known as the two main mechanisms of macrolide resistance8. The isolate had the erm(41) gene with a T28 sequevar and no rrl mutation acquisition.
The patient received oral clarithromycin and moxifloxacin, along with an initial 4-week course of intravenous amikacin and cefoxitin. During 4 weeks of initial antibiotic treatment, his symptoms and CT lesions were improved and sputum culture results were negative after 2, 3, and 4 weeks after treatment. Although he took oral clarithromycin and moxifloxacin for a total duration of 24 months, follow-up sputum cultures were persistently positive after 2 months after antibiotic treatment.
This is the first reported case of M. abscessus subsp. bolletii lung disease in Korea. Previous reports showed that patients with M. abscessus subsp. bolletii lung disease had mild compromised respiratory function, as was seen in the current report9,10. The reported number of patients with M. abscessus subsp. bolletii lung disease was less than 10 before this case9,10. The limited number of reports makes it difficult to characterize patients with M. abscessus subsp. bolletii lung disease.
The M. abscessus complex has undergone many taxonomic classification changes since its first description in 195311. The dramatic change in mycobacterial taxonomy came with the availability and reliability of DNA sequencing, including 16S rRNA, rpoB, and hsp65. Adekambi et al.12 suggested that the M. abscessus complex should be divided into three species, M. abscessus sensu stricto, M. massiliense, and M. bolletii, according to the results of rpoB sequencing. Recent evidence based on comparative analysis of whole genome sequences has suggested that M. abscessus can be divided into three subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii13. It is important to distinguish the three subspecies of M. abscessus complex, because of their differences in susceptibility to clarithromycin3. Commonly, M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii are inducible resistant, susceptible, and resistant to clarithromycin, respectively3.
The combination of macrolide, cefoxitin (or imipenem), and amikacin has been recommended as the first line antimicrobial drug treatment for M. abscessus lung disease14. However, the appropriate duration of intravenous antibiotic therapy or combination of oral antibiotics is not known. We used intravenous antibiotics for 4 weeks as previously reported cases of M. abscessus lung disease15, because of the phenotypic similarities between M. abscessus subsp. abscessus and M. abscessus subsp. bolletii. Although drug susceptibility testing showed in vitro resistance, clarithromycin and moxifloxacin were prescribed during outpatient treatment following the previous report3,15. We finally achieved improvement in his respiratory symptoms and prevented progression of radiological findings, although we were unable to achieve negative conversion of sputum AFB culture.
In summary, we report the first case of M. abscessus subsp. bolletii lung disease in Korea. M. abscessus subsp. bolletii lung disease should be considered when the pathogen is identified as M. abscessus complex by a non-sequencing method, such as PCR-RFLP, and has resistance to clarithromycin. In such cases, further molecular techniques may be needed to accurately identify the specific subspecies.
Figures and Tables
Figure 1
A 66-year-old male with bronchiectasis and nontuberculous mycobacterial lung disease caused by Mycobacterium abscessus subsp. bolletii. (A) Chest radiography revealed bilateral multifocal tram-track signs (black arrows), suggesting bronchiectasis. (B) A transverse chest computed tomography scan (3.0-mm-section thickness) revealed bilateral bronchiectasis (white arrows) in the middle right lobe and the lingular segment of the upper left lobe as well as multiple tree-in-bud appearances (black arrows), suggesting bronchiolitis in both lungs.
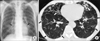
Figure 2
The phylogenetic position of isolate SMC-bol-002 and other species belong to the rapidly growing mycobacteria are based on rpoB sequences. This tree was constructed by using the neighbor-joining method and visualized with MEGA 5.0. The percentages at the nodes represent bootstrap levels which are supported by 1,000 re-sampled datasets. Scale bars indicate evolutionary distances in base substitutions per site. M.: Myobacterium.

Table 1
Antimicrobial drugs and MIC breakpoints

Drug susceptibility testing was performed by using a broth microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI)7.
MIC: minimum inhibitory concentration.
Acknowledgements
This work was supported by the Mid-career Researcher Program through a National Research Foundation grant funded by the Ministry of Education, Science and Technology (2011-0015546).
References
1. Koh WJ, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006; 129:341–348.
2. Macheras E, Roux AL, Bastian S, Leao SC, Palaci M, Sivadon-Tardy V, et al. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J Clin Microbiol. 2011; 49:491–499.
3. Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011; 183:405–410.
4. Koh WJ, Kwon OJ, Lee NY, Kook YH, Lee HK, Kim BJ. First case of disseminated Mycobacterium bolletii infection in a young adult patient. J Clin Microbiol. 2009; 47:3362–3366.
5. Lee H, Park HJ, Cho SN, Bai GH, Kim SJ. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000; 38:2966–2971.
6. Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, et al. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int J Syst Evol Microbiol. 2005; 55(Pt 4):1649–1656.
7. Clinical and Laboratory Standards Institute. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes: approved standard. 2nd ed. Wayne: CLSI;2011. Document no. M24-A2.
8. Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, et al. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother. 2011; 55:775–781.
9. Adekambi T, Drancourt M. Mycobacterium bolletii respiratory infections. Emerg Infect Dis. 2009; 15:302–305.
10. van Ingen J, de Zwaan R, Dekhuijzen RP, Boeree MJ, van Soolingen D. Clinical relevance of Mycobacterium chelonae-abscessus group isolation in 95 patients. J Infect. 2009; 59:324–331.
11. Moore M, Frerichs JB. An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region; report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J Invest Dermatol. 1953; 20:133–169.
12. Adekambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006; 56(Pt 1):133–143.
13. Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013; 381:1551–1560.
14. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007; 175:367–416.
15. Jeon K, Kwon OJ, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009; 180:896–902.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download