Abstract
We report a rare case of lung disease caused by Mycobacterium lentiflavum in a previously healthy woman. A 54-year-old woman was referred to our hospital due to chronic cough and sputum. A computed tomography scan of the chest revealed bilateral bronchiectasis with bronchiolitis in the right middle lobe and the lingular division of the left upper lobe. Nontuberculous mycobacteria were isolated twice from three expectorated sputum specimens. All isolates were identified as M. lentiflavum by multilocus sequence analysis based on rpoB, hsp65, and 16S rRNA fragments. To the best of our knowledge, this is the first documented case of M. lentiflavum lung disease in an immunocompetent adult in Korea.
Mycobacterium lentiflavum organisms are slow growing, tiny yellow nontuberculous mycobacteria (NTM) first identified in 19961. As with other NTM, M. lentiflavum has been isolated from soil and water samples around world2. It is a rare cause of human disease and has been reported to have clinical importance primarily in young children with cervical lymphadenitis and in immunocompromised patients3-5. Recently, however, a few cases of chronic pulmonary disease caused by M. lentiflavum in immunocompetent patients have been reported2,6-8.
Although the isolation of M. lentiflavum from clinical specimens has been reported in Korea, the infectious organisms seemed to be colonizers in the destructed lung due to previous tuberculosis9. Here, we report a very rare case of M. lentiflavum lung disease associated with bronchiectasis in an immunocompetent patient.
A 54-year-old woman was referred to our hospital due to chronic cough and sputum, which developed three years earlier. She had been a healthy, non-smoker up to this point, with the exception of pulmonary tuberculosis 10 years prior. Laboratory results, including immunoglobulin levels, were normal with the exception of an elevated erythrocyte sedimentation rate to 82 mm/hr. She had no risk factors for human immunodeficiency virus infection.
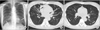
A chest radiography revealed bilateral multifocal tram-track signs. A computed tomography scan of the chest revealed bilateral bronchiectasis and bronchiolitis in the right middle lobe and the lingular division of the left upper lobe (Figure 1). There was no evidence of cystic fibrosis or other common causes of bronchiectasis. NTM were isolated twice from three consecutively expectorated sputum specimens.
Initial identification of the isolated NTM was unsuccessful using the polymerase chain reaction (PCR)-reverse blot hybridization assay (REBA) method based on the rpoB gene (REBA Myco-ID; M&D Inc., Wonju, Korea). No specific species could be assigned using this method due to a lack of reference sequences. Therefore, we used multilocus sequence analysis (MLSA) based on hsp65, rpoB, and 16S rRNA fragments to determine their taxonomic affiliations. Genomic DNA of the isolates was extracted using a commercially available kit (QIAamp DNA Mini kit; Qiagen, Hilden, Germany) and amplified using the appropriate pair of primers for each of the three genes. PCR conditions and primer sequences are described below. Partial amplification of the rpoB gene (723 bp) was performed using primers MycoF (5'-GGCAAGGTCACCCCGAAGGG-3') and MycoR (5'-AGCGGCTGCTGGGTGATCATC-3')10. Partial amplification of the hsp65 gene (439 bp) was performed using primers Tb11 (5'-ACCAACGATGGTGTGTCCAT-3') and Tb12 (5'-CTTGTCGAACCGCATACCCT-3')11. PCR conditions for these genes were as follows: 2 minutes at 95℃ followed by 35 cycles of 94℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 2 minutes, with a final extension step at 72℃ for 5 minutes. A 564 bp 5'-region of the 16S rRNA gene was amplified using primers 16MycF (5'-CGTGCTTAACACATGCAAGTCG-3') and 16MycR (5'-GTGAGATTTCACGAACAACGC-3')12. Conditions for 16S rRNA gene amplification were as follows: 2 minutes at 95℃ followed by 35 cycles of 94℃ for 30 seconds, 52℃ for 30 seconds, and 72℃ for 1 minute. The amplicons were cloned into the pGEM-TEasy vector (Promega, Madison, WI, USA), transformed into DH5α Escherichia coli and amplified (Invitrogen, Karlsruhe, Germany). Clones were sequenced using the ABI Prism d-Rhodamine dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA, USA). Sequencing reactions were recorded with an ABI Prism 3100 DNA sequencer following the standard protocol of the supplier (Applied Biosystems). The sequences were compared by referring to publically available sequence in the National Center for Biotechnology Information GenBank database13 and the Ribosomal Differentiation of Medical Micro-organisms (RIDOM) database14. Homogeneity values above 99% were considered significant. M. lentiflavum was subsequently identified as the most closely related species for which the sequences of the three genes were available in GenBank (accession nos. EU109300 for rpoB, AF547851 for hsp65, and GU142924 for 16S rRNA).
Five months after initial sputum cultures, the patient continued to have a mild cough and sputum. Additional sputum culture 4 months after the initial sputum examinations was negative. She was reluctant to receive long-term antibiotic therapy and refused further follow-up. Therefore, the long-term outcome of this patient is not available.
NTM are a well-known cause of chronic pulmonary disease. Among the most commonly isolated species are the M. avium complex, M. abscessus, and M. kansasii, but other NTM species have also been described as the cause of lung disease. To the best of our knowledge, this is the first documented case of M. lentiflavum lung disease in an immunocompetent adult in Korea, in which the etiologic organism was confirmed using MLSA based on rpoB, hsp65, and 16S rRNA fragments.
M. lentiflavum is a recently described slowly growing NTM1. It has been isolated primarily from lymph nodes of young children3, while isolations from other sites, including respiratory specimens, have been frequently described in immunocompromised patients4. M. lentiflavum lung disease in immunocompetent patients has rarely been described in the literature2,6-8, and the isolation of NTM always raises doubts about clinical significance. Our case confirms the potential pathogenicity of M. lentiflavum in chronic pulmonary disease.
However, the clinical significance of the isolation of M. lentiflavum in our case may be controversial, because of relatively short follow-up periods and small number of sputum acid-fast bacilli (AFB) analysis. As the diagnostic criteria for NTM lung disease are not proven pertinence to less common and low-virulence NTM, the patients with repeated isolations of low-virulence NTM require careful clinical evaluation and collection of multiple specimens for AFB analysis over time15. Considering that M. lentiflavum is known to have low-virulence, the patients may need to be followed with additional sputum examinations for longer time.
M. lentiflavum is phylogenetically related to other slow-growing NTM species such as M. avium, M. genavense, and M. simiae1. Traditional biochemical and/or phenotypic features of M. lentiflavum are not distinguishable from those of these closely related NTM species. Therefore, M. lentiflavum may have been previously misidentified as M. avium complex or M. simiae1,4. Accurate identification of the isolated NTM species is very important when initiating a treatment and selecting an antimicrobial regimen. However, M. lentiflavum cannot be identifiable by commercial kit for the identification of NTM species, such as PCR-restriction fragment length of polymorphism analysis or REBA Myco-ID based on the rpoB gene, because there were not included newly defined or infrequently species, especially for M. lentiflavum. Therefore, the prevalence of M. lentiflavum lung disease in Korea may be underestimated.
In conclusion, M. lentiflavum should be considered a possible etiologic pathogen of the nodular bronchiectatic form of NTM lung disease. We demonstrate here that diagnosis of M. lentiflavum can be verified by multilocus sequence analysis of rpoB, hsp65, and 16S rRNA fragments in clinical isolates.
Figures and Tables
Figure 1
A 54-year-old woman with bronchiectasis and nontuberculous mycobacterial lung disease caused by Mycobacterium lentiflavum. (A) A chest radiography reveals bilateral multifocal tram-track signs (white arrows). (B) A transverse chest computed tomography (CT) scan (2.5-mm-section thickness) on the level with the right inferior pulmonary vein reveals bilateral bronchiectasis (black arrowheads) in the right middle lobe and the lingular segment of the left upper lobe. (C) A chest CT scan obtained on the level with the superior segmental bronchus of the left lower lobe reveals bilateral bronchiolitis in both lungs (white arrowheads).
Acknowledgements
This work was supported by the Mid-career Researcher Program through a National Research Foundation grant funded by the Ministry of Education, Science and Technology (2011-0015546).
References
1. Springer B, Wu WK, Bodmer T, Haase G, Pfyffer GE, Kroppenstedt RM, et al. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996. 34:1100–1107.
2. Marshall HM, Carter R, Torbey MJ, Minion S, Tolson C, Sidjabat HE, et al. Mycobacterium lentiflavum in drinking water supplies, Australia. Emerg Infect Dis. 2011. 17:395–402.
3. Haase G, Kentrup H, Skopnik H, Springer B, Bottger EC. Mycobacterium lentiflavum: an etiologic agent of cervical lymphadenitis. Clin Infect Dis. 1997. 25:1245–1246.
4. Safdar A, Han XY. Mycobacterium lentiflavum, a recently identified slow-growing mycobacterial species: clinical significance in immunosuppressed cancer patients and summary of reported cases of infection. Eur J Clin Microbiol Infect Dis. 2005. 24:554–558.
5. Tortoli E, Mattei R, Russo C, Scarparo C. Mycobacterium lentiflavum, an emerging pathogen? J Infect. 2006. 52:e185–e187.
6. Tortoli E, Bartoloni A, Erba ML, Levre E, Lombardi N, Mantella A, et al. Human infections due to Mycobacterium lentiflavum. J Clin Microbiol. 2002. 40:728–729.
7. Molteni C, Gazzola L, Cesari M, Lombardi A, Salerno F, Tortoli E, et al. Mycobacterium lentiflavum infection in immunocompetent patient. Emerg Infect Dis. 2005. 11:119–122.
8. Shamaei M, Marjani M, Farnia P, Tabarsi P, Mansouri D. Human infections due to Mycobacterium lentiflavum: first report in Iran. Iran J Microbiol. 2010. 2:27–29.
9. Shin S, Yoon JH, Song SH, Kim EC. Isolation of Mycobacterium lentiflavum from a patient with a lung destroyed by tuberculosis. Korean J Lab Med. 2007. 27:124–127.
10. Adekambi T, Drancourt M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. 2004. 54(Pt 6):2095–2105.
11. Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993. 31:175–178.
12. Devulder G, Perouse de Montclos M, Flandrois JP. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int J Syst Evol Microbiol. 2005. 55(Pt 1):293–302.
13. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007. 35:D21–D25.
14. Harmsen D, Rothganger J, Frosch M, Albert J. RIDOM: Ribosomal Differentiation of Medical Micro-organisms Database. Nucleic Acids Res. 2002. 30:416–417.
15. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007. 175:367–416.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download