Abstract
Background
Mycobacterial infection is a problem throughout the world along with the increase of immunocompromised patients. For this reason, there have been many methods for faster and more accurate diagnosis. In this study, we evaluated several laboratory methods for mycobacterial infection.
Methods
From January to December 2009, 635 specimens were cultured with mycobacteria growth indicator tube (MGIT) and Ogawa media. Polymerase chain reaction (PCR) was performed with the AdvanSure tuberculosis (TB)/non-tuberculosis mycobacterium (NTM) real-time PCR Kit (LG Life Sciences, Seoul, Korea). The 69 samples showing positive culture results were identified with the AdvanSure Mycobacteria Genotyping Chip Kit (LG Life Science, Seoul, Korea).
Results
Sixty-nine (10.9%) out of 635 samples showed positive results for mycobacterial culture. Among the 635 samples, 64 were positive in MGIT, but only 42 were positive in Ogawa media. Of the 635 samples, 607 (95.6%) showed the same results between MGIT and Ogawa and the results of 579 (95.4%) were also consistent with the TB/NTM real-time PCR results. However, in the case of NTM, only one (1/24, 4.2%) was positive in PCR. In the Mycobacteria genotyping chip analysis, the most frequently identified NTM species in descending order were M. avium, M. intracellulare, M. chelonae and M. abscessus.
Conclusion
Culturing with a combination of MGIT and Ogawa is recommended to increase the recovery rate of mycobacteria. Although PCR missed a reasonable number of NTM, it is faster and usually gives results that concur with those from the culture. The appropriate combination of diagnostic methods with clinical correlation are necessary.
WHO reported that 9.4 million people (140 per 100,000 individuals) suffered from tuberculosis (TB) in 2009, representing a global health problem1. Infection with non-tuberculous mycobacterium (NTM) has been a problem throughout the world along with the spread of human immunodeficiency virus (HIV)2, and is also a problem in Korea among immunocompetent patients with chronic pulmonary diseases3.
There are several methods used to diagnose Mycobacterium tuberculosis (MTB) and NTM infection. Among them, acid-fast bacilli (AFB) smear is simple and able to produce results quickly but it has a low sensitivity and cannot differentiate between MTB and NTM. Mycobacterial culture has higher sensitivity and specificity than AFB smear but it takes a long time to verify the results as a long incubation time is required. To address this problem, liquid culture media was introduced. This can provide rapid and sensitive isolation of mycobacteria but the associated high contamination rate turned out to be a problem. Therefore, the combined use of liquid and solid media is recommended for quick and accurate verification2,4. TB-specific polymerase chain reaction (PCR) has been widely used to overcome the flaws of stains and culture tests but it is unable to detect NTM5. Recently, TB/NTM real-time PCR which can detect both TB and NTM was introduced and has been increasingly used5,6. Furthermore, the identification of NTM species using multiplex PCR or DNA chip became possible.
In this study, we evaluated the laboratory methods for assessing mycobacterial infection to provide baseline data for the establishment of a better diagnostic workflow for determining MTB and NTM infections.
From January to December 2009, mycobacterial culture and TB/NTM real-time PCR were performed with 635 samples from 511 patients. Among the samples, 405 were respiratory specimens (338 samples of sputum, 44 bronchial washings, and 23 pleural fluids). Others were CSF (36), body fluids (36), urines (31), tissues (93), wounds (28), whole blood (4) and bone marrow specimens (2).
For respiratory samples, NALC-NaOH solution (5% NaOH+0.5% NALC) was added to sample in container to liquify and decontaminate mucous sputum. The final volume was brought to 50 mL with phosphate buffer saline (PBS) and left at room temperature for 15 minutes after mixing. The solution was centrifuged at 3,000 g for 15 minutes at 4℃ and the supernatant was discarded. PBS (1~3 mL) was added after vortexing the sediment. CSF and other body fluids were processed with 5% NaOH without NALC and then processed in the same manner with respiratory samples. Processed samples were smeared and stained by Ziehl-Neelsen for AFB and read according to the criteria of Centers for Disease Control and Prevention (CDC) of America.
The processed samples (0.2 mL) were used to inoculate 3% Ogawa medium (Eiken, Tokyo, Japan) and were cultured for 8 weeks in an incubator at 35~37℃, 5~10% CO2. Interim report was provided at 4 weeks, if necessary. If colonies were grown on Ogawa media, colonies were stained for AFB.
0.5 mL of samples was used to inoculate MGIT medium (Becton Dickinson, Sparks, MD USA) after mixing with PANTA/Supplement according to the manufacturer's protocol. The tube was then incubated for 6 weeks in the BACTEC MGIT 960 system (Becton Dickinson). If fluorescence was detected in the tube, the test was considered positive. Samples from the tube were stained for AFB. If contaminants were observed, decontamination process was repeated.
Among samples showing AFB smear positive were tested using the AdvanSure Mycobacteria Genotyping Chip Kit (LG Life Science, Seoul, Korea) for mycobacterial identification. The AdvanSure Mycobacteria Genotyping Chip implements the method by employing the internal transcribed spacer (ITS) probe that exists between the 16S rRNA and 23S rRNA of mycobacteria. The chip was used according to the manufacturer's instruction. Hybridization was conducted after PCR and the results were analyzed and interpreted automatically with the LuxScan 10K microarray scanner (CapitalBio, Hong Kong, China).
DNA samples were extracted from clinical specimens and amplified using the AdvanSure TB/NTM real-time PCR kit following the manufacturer's protocol. SLAN real-time PCR detection system (LG Life Science, Seoul, Korea) was used to measure fluorescence formed during PCR process. A positive result was indicated when the cycle threshold (CT) value was less than 35 after observing signal formation of wavelength from each channel (FAM, HEX, and Cy5).
Among the 635 samples, 69 (10.9%) showed positive results in mycobacterial cultures. Of these 69 samples, 45 contained MTB and 24 contained NTM. In the Mycobacteria genotyping chip analysis, the most frequently identified NTM species in descending order were M. avium, M. intracellulare, M. chelonae and M. abscessus.
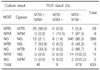
Among the whole samples, 64 were positive in MGIT, but only 42 samples were positive in Ogawa media (Table 1). Positivity of MGIT was higher than Ogawa media (MGIT 10.1%, Ogawa 6.6%).
MGIT detected 21 (26.9%) isolates of Mycobacteria from 78 AFB smear-positive samples and 43 (7.7%) isolates from 557 smear-negative samples. And Ogawa media detected 21 (26.9%) and 21 (3.8%) isolates, respectively. According to the degree of AFB smear (trace, 1+, 2+, 3+/4+), culture positivity was 8.3% (1/12), 23.1% (12/52), 63.6% (7/11) and 100% (3/3).
The contamination rate for 635 specimens noted with MGIT was 12.1% (77/635) while that with Ogawa media was 3.6% (23/635).
Of the 635 samples, 607 (95.6%) showed the same results in both MGIT and Ogawa (29 samples with TB, eight with NTM, and 570 with no growth). Additionally, among these 607 samples, 579 (95.4%) had results identical with those from TB/NTM real-time PCR (Table 2).
On the other hand, 32 (5.0%) of the 635 samples showed discordant results between MGIT and Ogawa. Among these, 13 were identified to contain TB by MGIT but no growth was observed in Ogawa, and seven of 13 were found positive by PCR. Three samples were identified to contain TB only by Ogawa and one of these was PCR-positive. 16 samples were identified as containing NTM by either media (14 in MGIT and two in Ogawa) but all of these were PCR-negative (Table 2).
Among 45 samples culture-positive for MTB, 36 (80.0%) were positive in TB/NTM real-time PCR. Among those 45 samples, 26 samples were negative in AFB smear. 19 of them were PCR-positive and 7 of them were PCR-negative. 19 samples were positive in AFB smear. 17 of them were PCR-positive and two of them were PCR-negative. Among 24 samples culture-positive for NTM, one (4.2%) was positive in TB/NTM real-time PCR. And that one was AFB smear-negative (Table 3).
The overall concordance rate between culture and TB/NTM real-time PCR was 93.1% for MGIT and 93.7% for Ogawa.
Among 69 samples showing positive results in MGIT or Ogawa, 45 samples (36 patients) were MTB. 34 of patients of them were diagnosed as TB. 24 samples (23 patients) were culture-positive for NTM. 13 patients had pulmonary diseases such as pneumonia and bronchiectasis and 4 of 13 patients were diagnosed as NTM pulmonary disease.
37 samples (33 patients) showed concordant results between MGIT and Ogawa media. Among them, 2 (6.1%) patients had taken anti-TB/NTM medications before. On the other hand, 32 samples (30 patients) showed discordant results between MGIT and Ogawa. 4 (13.3%) patients had taken anti-TB/NTM medications.
The positive rate of mycobacterial culture with combined use of liquid and solid media was 10.2%.
From the total 635 samples, positive identification rate of MGIT (10.1%) was higher than that of Ogawa (6.6%) (p=0.004, McNemar's test). MGIT culture for detecting mycobacteria has been found to be very useful. Kim et al.7 reported that among 99 culture-positive specimens, all were positive in MGIT, but only 64 were positive in Ogawa media. Therefore, MGIT is thought to have a superior recovery rate of mycobacteria compared to Ogawa media.
However, Yi et al.8 reported that out of 106 TB isolates, MGIT detected 95.3%, but MGIT plus Ogawa media detected 98.1%. Bae et al.2 also reported that recovery rates of mycobacteria using MGIT only, Ogawa only, and both media were 12.1%, 7.8%, and 13.3%, respectively. Therefore, Ogawa media should be used in combination with MGIT to increase the detection rate.
Additionally, MGIT and Ogawa media detected only 26.9% and 26.9% isolates of AFB smear-positive, respectively. Specimens showing trace or 1+ results from AFB smear usually showed culture-negative results. However, Bae et al.2 reported 67.9% of AFB smear-positive samples showed positive results in MGIT 960 system or Ogawa media. And Choi and Lee9 reported that all AFB smear positive samples resulted in cultures positivity.
In our study, the culture results from 579 (95.4%) of the 607 samples were identical to the ones from TB/NTM real-time PCR. Therefore, it seems that PCR is very useful for fast and accurate diagnosis of mycobacterial infections. However, for NTM detection, TB/NTM real-time PCR analysis showed a markedly low concordant rate with culture results. Among 24 samples found to have NTM in culture, only one (5.0%) showed a positive result for NTM by PCR.
In other studies evaluating Advansure TB/NTM real-time PCR, PCR positivity rates for NTM were 55.6% and 67.0% each studies among samples culture-positive for NTM6,10. The lower concordant rate in our study may be due to the small number of NTM-positive samples, so it will be necessary to study the NTM detection capacity of TB/NTM real-time PCR with a larger study group.
TB/NTM real-time PCR may not substitute for culture, especially for diagnosis of NTM disease. However, among no growth samples in cultures, 20 (MTB:12, NTM:8) showed PCR-positive. And among these cases, nine were really TB patients and two patients had NTM pulmonary disease. As stated above, TB/NTM real-time PCR cannot be used only, but it will be useful to diagnose TB or NTM disease adjuctly.
NTM infection rates have been increasing. Although NTM is normally present in natural environments such as water and soil, it was reported that approximately 40~50% of patients in the Unites States and 10~17% of patients in Asia who were found to be infected with NTM by culture had NTM-related pulmonary diseases11-15. Moreover, the fact that NTM has high resistant rates against multiple antibiotics and its treatment can be different from TB underscores the need for making more accurate differential diagnosis between TB and NTM infections3.
With the advancement of laboratory medicine, diagnostic methods for the detection of mycobacterial infection have also been remarkably developed. At present, typical method such as AFB smear to molecular-based methods are used together according to laboratory circumstances.
When specimens are referred to our laboratory, AFB smear using the Ziehl-Neelsen method is performed with decontaminated samples. Next, they are used to inoculate MGIT and Ogawa media. If the samples show positive signals from MGIT or visible colonies on Ogawa media, AFB smear is performed again using samples from MGIT and Ogawa. If AFB smear is also positive, Mycobacteria genotyping chip analysis is performed for mycobacteria species identification. TB/NTM real-time PCR is also performed with cultures simultaneously if it is thought to be necessary for diagnosis.
As shown in many studies, every laboratory method has specific pros and cons. Therefore, the appropriate combination and regular assessment of diagnostic methods with clinical correlation are necessary to improve the reliability of laboratory analysis technique.
Figures and Tables
References
1. Global health observatory (GHO): tuberculosis. World Health Organization (WHO). c2011. cited 2011 Oct 7. Geneva, Switzerland: WHO;Available from: http://www.who.int/mediacentre/factsheets/fs104/en/.
2. Bae E, Im JH, Kim SW, Yoon NS, Sung H, Kim MN, et al. Evaluation of combination of BACTEC mycobacteria growth indicator tube 960 system and Ogawa media for Mycobacterial culture. Korean J Lab Med. 2008. 28:299–306.
3. Lee HW, Kim MN, Shim TS, Bai GH, Pai CH. Nontuberculous mycobacterial pulmonary infection in immunocompetent patients. Tuberc Respir Dis. 2002. 53:173–182.
4. Joung US, Jeong J, Lee SH, Kim SR. Comparison of mycobacterial culture by mycobacterium growth indicator tube and Ogawa media. Korean J Clin Microbiol. 2004. 7:135–138.
5. Yun EY, Cho SH, Go SI, Baek JH, Kim YE, Ma JE, et al. Usefulness of real-time PCR to detect mycobacterium tuberculosis and nontuberculous mycobacteria. Tuberc Respir Dis. 2010. 69:250–255.
6. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. Evaluation of the performances of advanSure TB/NTM real time PCR kit for detection of mycobacteria in respiratory specimens. Korean J Lab Med. 2008. 28:34–38.
7. Kim YS, Jo YH, Lee HJ, Suh JT, Lee YJ. Comparison of the MGIT (mycobacteria growth indicator tube) with Ogawa media for recovery of mycobacteria. Korean J Clin Microbiol. 2001. 4:58–61.
8. Yi JY, Kim JP, Shin JH, Suh SP, Ryang DW. Detection of mycobacterium tuberculosis using BACTEC mycobacteria growth indicator tube (MGIT) 960 system: comparison with BACTEC 460 TB system and Ogawa media. Korean J Clin Pathol. 2000. 20:384–391.
9. Choi YM, Lee MH. Evaluation of the BACTEC MGIT 960 system for the recovery of mycobacteria. Korean J Clin Pathol. 2000. 20:56–61.
10. Jung CL, Kim MK, Seo DC, Lee MA. Clinical usefulness of real-time PCR and amplicor MTB PCR assays for diagnosis of tuberculosis. Korean J Clin Microbiol. 2008. 11:29–33.
11. Lee JK, Kwon HY, Kwon JK, Lee HJ, Lee DW, Lee YJ, et al. Recovery rate of nontuberculous mycobacteria and the clinical course of nontuberculous mycobacterial pulmonary disease at a secondary hospital. Tuberc Respir Dis. 2009. 67:199–204.
12. Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, et al. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med. 2010. 182:977–982.
13. O'Brien RJ, Geiter LJ, Snider DE Jr. The epidemiology of nontuberculous mycobacterial diseases in the United States. Results from a national survey. Am Rev Respir Dis. 1987. 135:1007–1014.
14. Sakatani M. Nontuberculous mycobacteriosis; the present status of epidemiology and clinical studies. Kekkaku. 1999. 74:377–384.
15. Hosker HS, Lam CW, Ng TK, Ma HK, Chan SL. The prevalence and clinical significance of pulmonary infection due to non-tuberculous mycobacteria in Hong Kong. Respir Med. 1995. 89:3–8.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download