Abstract
Background
Transcription factor FOXP3 characterizes the thymically derived regulatory T cells. FOXP3 is expressed by cancer cell itself and FOXP3 expression was induced by TGF-β treatment in pancreatic cancer cell line. However, the expression of FOXP3 expression is not well known in patients with lung cancer. This study was conducted to investigate the expression of FOXP3 in patients with lung cancer and to investigate the regulation of FOXP3 expression by the treatment of TGF-β and DNA methyltransferase inhibitor in lung cancer cell lines.
Methods
FOXP3 expression in the tissue of patients with resected non-small cell lung cancer (NSCLC) was evaluated by immunohistochemistry. The regulation of FOXP3 expression was investigated by Western blot and RT-PCR after lung cancer cell lines were stimulated with TGF-β1 and TGF-β2. The regulation of FOXP3 expression was also investigated by RT-PCR and flow cytometry after lung cancer cell lines were treated with DNA methyltransferase inhibitor (5-AZA-dC).
Results
FOXP3 expression was confirmed in 27% of patients with NSCLC. In NCI-H460 cell line, TGF-β2 decreased FOXP3 mRNA and protein expressions. In A549 cell line, both TGF-β1 and TGF-β2 decreased FOXP3 mRNA and protein expressions. 5-AZA-dC increased FOXP3 mRNA expression in NCI-H460 and A549 cell lines. Moreover, 5-AZA-dC increased intracellular FOXP3 protein expression in A549 cell lines.
FOXP3 is a member of Forkhead transcription factor family. It has been known to be a specific marker of regulatory T cells (Treg)1,2. The expression of FOXP3 in tumor cells themselves has been shown first in hematologic malignancy such as adult T-cell leukemia/lymphoma (ATLL). Recently, the expression of FOXP3 in patients with tumor tissues of breast cancer, pancreas cancer, prostate cancer and other solid tumor has been reported3-7. In studies performed on cell lines of melanoma, colorectal cancer, lung cancer, prostate cancer, and breast cancer, applied flow cytometry and qRT-PCR, the expression of FOXP3 within tumor cells has been reported8,9. Recently, the expression of FOXP3 within cancer cells in tumor tissues of pancreas cancer and breast cancer patients was confirmed by immunohistochemical staining. In addition, it has been reported that the expression of FOXP3 within tumor cells is associated with tumor progression and metastasis, and thus it is a poor prognostic factor4,5. Furthermore, when pancreas cancer cells expressing FOXP3 derived from actual patients were cultured with naïve T cells, the proliferation of T cells was suppressed, and tumor cells expressing FOXP3 mediated immune suppression reactions similar to Treg, and thus the hypothesis that the expression of FOXP3 in tumor cells is a new mechanism of tumor cells evading immunity and it may be associated with poor prognosis has been proposed4. In addition, it has been reported that the expression of FOXP3 in breast cancer tissues is associated with distant metastasis and it is a poor prognostic factor5.
In regard to the mechanism of the regulation of the expression of FOXP3 in tumor tissues, it has been reported that in pancreas cancer cell lines, similar to Treg, the expression of FOXP3 is induced by TGF-β. However, the role of TGF-β in the expression of FOXP3 in other cancer cell lines has not been shown yet4. It has been reported that in CD4+ CD25- T cells, FOXP3 promoter is demethylated by TGF-β treatment resulting in the increase of the expression of FOXP3, hence, a mechanism that controls the expression of FOXP3 by TGF-β treatments without the interaction with TGF-β receptor has been also shown10. Therefore, in tumor cell lines including lung cancer, it is possible that the expression of FOXP3 is regulated by the TGF-β pathway as well as the suppression of DNA methylation.
Until now, it has not been reported whether FOXP3 is expressed in the patients with lung cancer tissues. Inferring from the result of other previous studies on solid tumors, it appears that the possibility of FOXP3 being expressed in lung cancer tissues and the expression being a poor prognostic factor is high. Lung cancer responds poorly to chemotherapy and its prognosis is poor, and thus it is an important task to seek other treatment methods including immunotherapy. Since FOXP3 expression is associated with immune evasion and distant metastasis in previous studies, the regulation of FOXP3 expression in cancer cells may be applied to the treatment of lung cancer patients.
The purpose of this study was to examine whether FOXP3 is expressed in lung cancer tissues by immunohistochemical staining, and to evaluate the modulation of the expression of FOXP3 in response to TGF-β treatment as well as the treatment with DNA methyltransferase inhibitor.
Selected among patients newly diagnosed as non-small cell lung cancer at the Samsung Medical Center from 1994 to 1996, the subjects were patients who underwent radical resection surgery. For immunohistochemical staining, tissue microarray blocks were used. For the enzyme-induced epitope retrieval, proteinase K was used. As primary antibody, monoclonal anti-FOXP3 antibody (Abcam, Cambridge, MA, USA) was used. The samples were treated with the ABC reagent (Vectastatin ABC kit; Vector Laboratories, Inc., Burlingame, CA, USA), color reactions were induced by the use of 3,3-diaminobenzidine (DAB), and examined. The study was performed after obtaining the approval from the institutional review board of the Samsung Medical Center.
According to previous studies, it has been shown that A549 cells (adenocarcinoma cell line) and NCI-H460 cells (large cell carcinoma cell line) express FOXP38. Thus, in our study, for experiments assessing the regulation of the expression of FOXP3, A549 and NCI-H460 cell lines were used. The expression of FOXP3 in NCI-H146 cells (small cell carcinoma cell line) was examined but it was not detected, and thus the cells were not used in experiments that examined the mechanism of the regulation of the expression of FOXP3. Tumor cell lines were cultured in RPMI 1640 culture medium (supplemented with 10% fetal calf serum, 2 mmol/L glutamine and 1 mmol/L sodium pyruvate), in a constant temperature and humidity incubator (37℃ and 5% CO2).
Total RNA of tumor cell lines was isolated using the TRIzol Plus RNA Purification System (Invitrogen, Grand Island, NY, USA). For reverse transcriptase-polymerase chain reaction (RT-PCR), Superscript II reverse transcriptase (Gibco BRL, NY, USA) was used. The used FOXP3 primer sequence is as follows4. FOXP3 sense: 5'CACAACATGCGACCCCCTTTCACC 3'; FOXP3 antisense: 5'AGGTTGTGGCGGATGGCGTTCTTC 3'. As the internal control, actin was used, and the primer sequences are as follows. Actin sense: 5'CGCGGACTATGACTT-AGTTG 3; actin antisense: 5'AAACAACAATGTGCAATCAA3'. PCR was performed in total 20 µL reaction solution containing 100 ng templates, 1.4 pmol/µL primer, 0.25 mM dNTP, 2.0 mM MgCl2, and 0.025 U/µL Taq polymerase. As PCR for FOXP3, the initial denaturation at 95℃ for 10 minutes was performed, and subsequently, 35 cycles of the reaction at 95℃ for 15 minutes, 60℃ for 15 seconds, and 72℃ for 15 seconds were performed. As PCR for actin, the initial denaturation 95℃ for 10 minutes was performed. Subsequently, 25 cycles of the reactions at 95℃ for 15 seconds, 60℃ for 15 seconds, and 72℃ for 15 seconds were performed. After the completion of the reactions, PCR products were electrophoresed on 1% agarose gel, stained with ethidium bromide and examined.
1) Western blot: Cultured tumor cells were collected, and proteins were isolated by the use of whole cell lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.02% Na-azide, 1% NP-40, 200 µM PMSF). 15~20 µg proteins were electrophoresed on 12.5% SDS-PAGE gel, and electrotransfer was performed for 90 minutes. The samples were reacted with FOXP3 antibody (PCH101 clone; eBioscience, Hatfield, UK) diluted to 1:100 at 4℃ overnight. Subsequently, the samples were reacted with the secondary horseradish peroxidase conjugated antibody, and the expression of FOXP3 was assessed by the application of enhanced chemiluminescence.
2) Flow cytometry: To assess the intracellular expression of FOXP3 protein, Alexa FluorR 488 anti-human FOXP3 antibody (BioLegend, San Diego, CA, USA), Alexa FluorR 488 mouse IgG1 (BioLegend), k isotype control (BioLegend), and FOXP3 Fix/Perm buffer set (BioLegend) were used. In each experiment, approximately 2×105 tumor cells were used, and approximately 2×104 cells (10%) were analyzed. Experiments were performed by the FACSCalibur (BD Biosciences, Sparks, MD, USA) installed Cellquest Pro software. For data analysis, the FlowJo 7.5 (Tree Star, Ashland, Oregon, USA) was used.
A549 cells and NCI-H460 cells were stimulated with 5 ng/mL each TGF-β1 (R&D Systems Inc., USA) and TGF-β2 (R&D Systems Inc., USA) for 48 hours. After 48 hours incubation, tumor cells of the TGF-β1 treatment group, TGF-β2 treatment group and the untreated group were harvested, and for the assessment of the expression of protein, Western blot was performed. As the positive control, FOXP3 293T cell transient overexpression lysate (Novus Biologicals, USA) was used. As the negative control, BEAS-2B cell line was used. In NCI-H460 cell line, the expression of FOXP3 mRNA according to the concentration of TGF-β at various times was assessed. In A549 cell line, cells were stimulated with 5 ng/mL TGF-β1 or TGF-β2 for 48 hours, and the expression of mRNA was assessed by RT-PCR.
In A549 cells and NCI-H460 cells, the modulation of the expression of FOXP3 mRNA in response to the DNA methyltransferase inhibitor (5-AZA-dC, Sigma-Aldrich, USA) was examined by RT-PCR. The modulation of the expression of FOXP3 mRNA according to the concentration of 5-AZA-dC treatment (0, 1 µM and 10 µM) as well as the treatment time (24-, 48-, 72- and 96 hours) was evaluated. The modulation of the expression of FOXP3 protein during 48 hours of the treatment with different concentrations of 5-AZA-dC (0, 0.1 µM, 1 µM and 10 µM) was examined by flow cytometry analysis.
Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. 100 µL cell suspension containing 1×104 tumor cells were seeded in each well of 96-well plates. Afterward, cells were treated with 5-AZA-dC at given concentrations and times, and subsequently, 20 µL MTT was added to each well. The final concentration was 5 mg/mL. To allow MTT to react with living cells and form formazan crystals, cells were incubated with MTT for 4 hours, and then treated with dimethylsulfoxide (DMSO) at 37℃ for 10 minutes. The optical density of each well was measured by a microplate reader at 495 nm. Cell viability was calculated by the following formula. Cell viability (%)=(absorbance of the treated wells-absorbance of the blank control wells)/(absorbance of the negative control wells-absorbance of the blank control wells)×100%11. All MTT experiments were performed in triplicates.
1) The expression of FOXP3 protein in lung cancer tissues: The expression of FOXP3 protein in tumor cells obtained from lung cancer patient tissues was assessed by immunohistochemical staining (Figure 1A). Of total 96 study subject patients, the expression of FOXP3 was confirmed in 27 patients (28.1%). The expression of FOXP3 was not different according to histological types.
2) The expression of FOXP3 mRNA and protein in lung cancer cell lines: In NCI-H460 cells and A549 cells, the expression of FOXP3 mRNA (Figure 1B) and protein (Figure 1C) was detected. Nevertheless, in NCI-H146 cells, the expression of neither FOXP3 mRNA (Figure 1B) nor protein (Figure 1C) was detected.
NCI-H460 cell lines were treated for 48 hours with 0.1-, 1-, 5- or 10 ng/mL TGF-β1 (Figure 2A) or TGF-β 2 (Figure 2B), and the expression of FOXP3 mRNA was compared with the control group. Treated with 1-, 5- or 10 ng/mL TGF-β1, the expression of FOXP3 mRNA was increased by 2.0, 1.8 or 2.6 times, respectively (Figure 2A). In the group treated with 10 ng/mL TGF-β 2, the expression of FOXP3 was decreased by 0.4 times (Figure 2B). NCI-H460 cell line was treated with 5 ng/mL TGF-β1 (Figure 2C) or TGF-β2 (Figure 2D) for various times and the expression of FOXP3 mRNA was examined. After 6 hours incubation with TGF-β1, the expression of FOXP3 mRNA was increased by 1.5 times. Nonetheless, it showed a trend to be decreased afterward (Figure 2C). In response to TGF-β2 stimulation, the expression of FOXP3 mRNA was not greatly modulated (Figure 2D). When NCI-H460 cells were treated for 48 hours with 5 ng/mL TGF-β1 or 5 ng/mL TGF-β2, in comparison with the control group, a trend of the reduced expression of FOXP3 protein was shown (Figure 2E). The results of 3 independent experiments showed that in response to TGF-β1 or TGF-β2 stimulation, in comparison with the control group, the reduction of the expression of FOXP3 protein by average 0.4 times or 0.5 times, respectively, was shown (Figure 2F).
In A549 cell line, to examine the effect of TGF-β1 and TGF-β2 on the expression of FOXP3 mRNA and protein, after the stimulation with TGF-β1 and TGF-β2, the regulation of the expression of FOXP3 was evaluated. In A549 cell line, after the treatment with 5 ng/mL TGF-β1 or 5 ng/mL TGF-β2 for 48 hours, the expression of FOXP3 mRNA was decreased by 0.2 times and 0.6 times, respectively (Figure 3A). In A549 cell line, after the treatment with 5 ng/mL TGF-β1 or 5 ng/mL TGF-β2 for 48 hours, a trend of the decrease of the expression of FOXP3 protein was shown (Figure 3B). The results of 3 independent experiments show that by the stimulation with TGF-β1 or TGF-β2, in comparison with the control group, the expression of FOXP3 protein was reduced by average 0.6 times (Figure 3C).
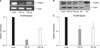
The regulation of the expression of FOXP3 mRNA in response to the treatment with 5-AZA-dC in lung cancer cell lines was examined. In NCI-H460 cell line, the expression of FOXP3 mRNA according to the concentration of 5-AZA-dC and the treatment time was examined (Figure 4A). After 24 hours, treated with 1 or 10 µM concentration, the expression of FOXP3 mRNA was increased by 3.4 times or 5.6 times, respectively, in comparison with the control group. Afterward, it showed a trend to be decreased (Figure 4A). In A549 cell line, similarly, the expression of FOXP3 mRNA according to the concentration and time of 5-AZA-dC treatment was assessed (Figure 4B). In A549 cell line, after 24 hours of stimulation with 1 µM 5-AZA-dC, the expression of FOXP3 mRNA was increased by 2.5 times in comparison with the control group. After 48 hours treatment with 1 µM or 10 µM 5-AZA-dC treatment, it was increased by 2.7 and 3.5 times, respectively, in comparison with the control group. The change of the cell viability of lung cancer cell lines in response to 5-AZA-dC treatment was also examined. NCI-H460 cell line and A549 cell line were treated with 1 or 10 µM 5-AZA-dC for 24, 48, 72 or 96 hours. In both NCI-H460 cell line (Figure 4C) and A549 cell line (Figure 4D), significant reduction of cell viability in comparison with the control group was not detected. It thus was confirmed that the treatments with up to 10 µM concentration of 5-AZA-dC for up to 96 hours did not mediate effects on cell viability.
It was examined whether the intracellular expression of FOXP3 in A549 cell line was modulated in response to 5-AZA-dC treatment by flow cytometry. A549 cell line was treated for 48 hours with 0.1-, 1- or 10 µM 5-AZA-dC, and the intracellular expression of FOXP3 was evaluated. It was observed that treated with 1 or 10 µM 5-AZA-dC, the intracellular expression of FOXP3 was elevated in comparison with the control group (Figure 5).
In our study, it was confirmed that FOXP3 protein was expressed within actual lung cancer tissue of patients. It was also observed that FOXP3 mRNA and protein were expressed in NCI-H460 and A549 lung cancer cell lines. In NCI-H460 cells, TGF-β1 elevated the expression of FOXP3 mRNA, nonetheless, the expression of FOXP3 protein rather showed a tendency to be reduced, and thus a concurrent trend was not shown. In NCI-H460 cells, TGF-β2 reduced the expression of FOXP3 mRNA and reduced the expression of FOXP3 protein. In A549 cells, both TGF-β1 and TGF-β2 reduced the expression of FOXP3 mRNA, and it also showed a tendency to reduce the expression of FOXP3 protein. On the other hand, in response to the treatment with the DNA methyltransferase inhibitor, FOXP3 mRNA was increased in both NCI-H460 cells and A549 cells. Similarly, the intracellular expression of FOXP3 was also increased in response to 5-AZA-dC treatment in A549 cells. Therefore, it appears that in the expression of FOXP3 in tumor cells, epigenetic mechanisms such as the modification of DNA methylation may play an important role.
It has been shown that FOXP3 is a transcription factor expressed selectively in Treg. Recently, in studies conducted on melanoma, colorectal cancer, lung cancer, prostate cancer, breast cancer, and other various cancer cell lines applying flow cytometry and qRT-PCT, the expression of FOXP3 in tumor cells has been reported8,9. In studies examined the expression of FOXP3 in A549, NCI-H460 and CaLu-6 lung cancer cell lines by flow cytometry, the mean fluorescence intensity was higher than the control group fibroblasts8. In lung cancer cell lines such as CaLu-1, CaLu-6, GILI, ONET, SK-LU-1, NCI-H441, NCI-H460, NCI-H596 and NCI-H661 cells, according to studies examined the expression of FOXP3 mRNA by qRT-PCR, the expression of FOXP3 mRNA was elevated in GILI, NCI-H460 and NCI-H661 in comparison with the control fibroblasts9. Karanikas et al.9 confirmed that in a lung adenocarcinoma cell line (GILI), FOXP3 protein was shown to be expressed in the nucleus as well as the cytoplasm by immunohistochemical staining. In our study, similarly, the expression of FOXP3 mRNA and protein in A549 as well as NCI-H460 cells was confirmed, and the expression of FOXP3 in the nucleus and cytoplasm of tumor cells of actual lung cancer tissues of patients was confirmed, which is in agreement with the result of other studies.
Until now, comprehensive studies on the expression of FOXP3 in tumor cell lines including lung cancer have not been conducted. Hinz et al. have reported that in a pancreatic ductal adenocarcinoma cell line as well as tumor cells obtained from pancreas cancer patient tissues, FOXP3 was expressed, the expression of FOXP3 was not induced by the stimulation with 5 ng/mL TGF-β1 for 48 hours, and it was induced only by the stimulation with 5 ng/mL TGF-β2 for 48 hours4. However, in our study, both NCI-H460 and A549 lung cancer cell line were treated with diverse concentrations of TGF-β1 or TGF-β2 for varying times, the expression was evaluated, and the results different from previous studies on pancreatic cancer cell lines were obtained. In our study, in response to the stimulation of NCI-H460 cells with TGF-β2, the expression of both FOXP3 mRNA and protein was decreased. In addition, in A549 cells, in response to the stimulation with TGF-β1 or TGF-β2, the expression of both FOXP3 mRNA and protein was reduced. Such results suggest the possibility of the different effect of TGF-β1 and TGF-β2 on the expression of FOXP3. It has been shown that TGF-β plays a role of suppressing the development of tumors by maintaining the homeostasis of normal epithelial cells during the early stage of the development of tumors, and at the late phase of the development of tumors, it suppresses the immunity in tumor tissues and accelerates tumor metastasis, and thus it could play contradictory roles that contribute to tumor progression12-14. Therefore, it is thought that the regulation of the expression of FOXP3 by TGF-β in tumor cell lines may show different results depending on the stage of tumor and the type of tumors. In regard to the regulation of FOXP3 by TGF-β, additional studies on diverse tumor cell lines should be performed in the future.
The role of epigenetic regulation in the expression of FOXP3 in tumor cells has not been reported previously. In our study, it was observed that in NCI-H460 and A549 lung cancer cell line, 5-AZA-dC increased the expression of FOXP3 mRNA. In addition, the intracellular expression of FOXP3 protein assessed by flow cytometry showed a tendency to be increased in response to 5-AZA-dC treatment. According to previous studies, it has been reported that in pancreas cancer and breast cancer patients, the expression of FOXP3 in tumor cells was associated with tumor progression and metastasis, and thus it is a poor prognostic factor4,5. Therefore, it is possible that the elevation of the expression of FOXP3 in tumor cells facilitates tumor cells to evade immunity and distant metastasis, and thus shortens the survival period of patients. It is thought that if the mechanism that could regulate the expression of FOXP3 in tumor cells is characterized by additional studies, it may exert effects on the treatment as well as prognosis of patients with lung cancer.
Summarizing the results, in our study, the expression of FOXP3 in tumor cells of lung cancer tissues of patients was confirmed. In NCI-H460 and A549 cell lines, TGF-β2 reduced the expression of FOXP3, the suppression of DNA methylation increased the expression of FOXP3, and thus the possibility that TGF-β and DNA methylation may play an important role in the expression of FOXP3 in lung cancer cell lines was observed. It appears that for the comprehensive understanding of the association of the expression of FOXP3, survival and prognosis in lung cancer, studies examining whether other epigenetic mechanisms such as histone modification in tumor cell lines are involved in the regulation of the expression of FOXP3 are required in the future.
Figures and Tables
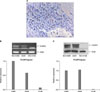 | Figure 1FOXP3 expression in the tissue of patient with lung cancer and lung cancer cell lines. (A) Immunohistochemical staining of paraffin-embedded non-small cell lung carcinoma tissue revealed FOXP3 expression in lung carcinoma cells (H&E stain, ×400). (B) FOXP3 mRNA expression was confirmed in NCI-H460 and A549 cell lines. (C) FOXP3 protein expression was confirmed in NCI-H460 and A549 cell lines. |
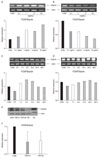 | Figure 2FOXP3 mRNA and protein expression after TGF-β treatment of NCI-H460 cell lines. The expression of FOXP3 mRNA after stimulation with TGF-β1 (A) and TGF-β2 (B) with different concentrations in NCI-H460 cell lines for 48 hours. The expression of FOXP3 mRNA after stimulation with 5 ng/mL TGF-β1 (C) and 5 ng/mL TGF-β2 (D) in NCI-H460 cell lines for given times. (E) The expression of FOXP3 protein after stimulation with 5 ng/mL TGF-β1 and 5 ng/mL TGF-β2 for 48 hours in NCI-H460 cell lines. (F) Mean relative expression of FOXP3 expression after stimulation with 5 ng/mL TGF-β1 and 5 ng/mL TGF-β2 for 48 hours from 3 independent experiments of NCI-H460 cell lines. CON: no treatment; NC: negative control (BEAS-2B cell line); PC: positive control (FOXP3 293T cell transient overexpression lysate). |
 | Figure 3FOXP3 mRNA and protein expression after TGF-β treatment of A549 cell lines. (A) The expression of FOXP3 mRNA decreased after stimulation with 5 ng/mL TGF-β1 and 5 ng/mL TGF-β2 in A549 cell lines for 48 hours. (B) The expression of FOXP3 protein after stimulation with 5 ng/mL TGF-β1 and 5 ng/mL TGF-β2 in A549 cell lines for 48 hours. (C) Mean relative expression of FOXP3 expression after stimulation with 5 ng/mL TGF-β1 and 5 ng/mL TGF-β2 for 48 hours from 3 independent experiments of A549 cell lines. CON: no treatment; NC: negative control (BEAS-2B cell line); PC: positive control (FOXP3 293T cell transient overexpression lysate). |
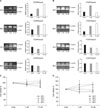 | Figure 4FOXP3 mRNA expression after 5-AZA-dC treatment of NCI-H460 and A549 cell lines. (A) The expression of FOXP3 mRNA after treatment with 5-AZA-dC with given concentrations for 24-, 48-, 72-, and 96 hours in NCI-H460 cell lines. (B) The expression of FOXP3 mRNA after treatment with 5-AZA-dC with given concentrations for 24-, 48-, 72-, and 96 hours in A549 cell lines. 5-AZA-dC treatment did not have significant effects on cell viability of NCI-H460 (C) and A549 (D) cell lines. NCI-H460 and A549 cell lines were incubated with 5-AZA-dC (1 and 10 µM) for 24-, 48-, 72-, and 96 hours. Cell viability was assessed by the MTT assay. CON: control. |
 | Figure 55-AZA-dC treatment and intracellular FOXP3 expression in A549 cell lines. Intracellular FOXP3 expression after treatment of 5-AZA-dC with given concentrations for 48 hours in A549 cell lines. The cells were stained using Alexa FluorR 488 anti-human FOXP3 antibody (solid line) or isotype-matched control antibody (dotted line) and analyzed by flow cytometry. |
Acknowledgements
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A090909).
References
1. Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005. 22:329–341.
2. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003. 299:1057–1061.
3. Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W, et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007. 129:1275–1286.
4. Hinz S, Pagerols-Raluy L, Oberg HH, Ammerpohl O, Grüssel S, Sipos B, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007. 67:8344–8350.
5. Merlo A, Casalini P, Carcangiu ML, Malventano C, Triulzi T, Mènard S, et al. FOXP3 expression and overall survival in breast cancer. J Clin Oncol. 2009. 27:1746–1752.
6. Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007. 117:3765–3773.
7. Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009. 16:336–346.
8. Ebert LM, Tan BS, Browning J, Svobodova S, Russell SE, Kirkpatrick N, et al. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008. 68:3001–3009.
9. Karanikas V, Speletas M, Zamanakou M, Kalala F, Loules G, Kerenidi T, et al. Foxp3 expression in human cancer cells. J Transl Med. 2008. 6:19.
10. Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007. 204:1543–1551.
11. Cui DD, Huang Y, Mao SH, Chen SC, Qiu M, Ji LL, et al. Synergistic antitumor effect of TRAIL and adriamycin on the human breast cancer cell line MCF-7. Braz J Med Biol Res. 2009. 42:854–862.
12. Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999. 13:804–816.
13. Massagué J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006. 580:2811–2820.
14. Park C, Kim WS, Choi Y, Kim H, Park K. Effects of transforming growth factor beta (TGF-beta) receptor on lung carcinogenesis. Lung Cancer. 2002. 38:143–147.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download