Abstract
Background :
The aberrant promoter hypermethylation of p16INK4a, as a tumor suppressor gene, is contributory factor to non-small cell lung cancer(NSCLC). However, its potential diagnostic impact of lung cancer is unclear. This study measured the level of p16INK4a promoter hypermethylation in the sputum and blood, and compared this with the level measured in the tissue obtained from NSCLC and pulmonary inflammation.
Methods :
Of the patients who visited the Our Lady of Mercy Hospital in Incheon, Korea for an evaluation of a lung mass and underwent blood, sputum, and tissue tests, 23patients (18 NSCLC, 5 pulmonary inflammation) were enrolled in this study. DNA was extracted from each sample and the level of p16INK4amethylation was determined using methylation-specific polymerase chain reaction.
Results :
p16INK4a methylation of the blood was observed in 88.9% (16 of 18) and 20.0% (1 of 5) of NSCLC and from pulmonary inflammation samples, respectively (P=0.008). Methylation of the sputum was observed in 83.3% (10 of 12) 80.0% (4 of 5) of NSCLC and pulmonary inflammation samples, respectively (P=1.00). Among the 8 NSCLC tissue samples, methylation changes were detected in 75.0% of samples (6 cases). Four out of seven tissue samples (57.1%) showed concordance, being methylated in both the blood and sputum.
Conclusions :
There was a higher level of p16INK4a methylation of the blood from NSCLC patients than from pulmonary inflammation. The tissue showed a high concordance with the blood in the NSCLC samples. These findings suggest that p16INK4a promoter hypermethylation of the blood can used to discriminate between NSCLC and pulmonary inflammation.
Go to : 
REFERENCES
1.Greenlee RT., Murray T., Bolden S., Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000. 50:7–33.

2.통계청. 2003년사망원인통계결과. www.nso.go.kr. ).2004.
3.Eddy DM. Screening for lung cancer. Ann Intern Med. 1989. 111:232–7.
4.Martin J., Ginsberg RJ., Venkatraman ES., Bains MS., Downey RJ., Korst RJ, et al. Long-term results of combined–modality therapy in resectable non-small –cell lung cancer. J Clin Oncol. 2002. 20:1989–95.
5.Weiss W., Boucot KR. The Philadelphia Pulmonary Neoplasm Research Project. Early roentgenographic appearance of bronchogenic carcinoma. Arch Intern Med. 1974. 134:306–11.

6.An evaluation of radiologic and cytologic screening for the early detection of lung cancer: a cooperative pilot study of the American Cancer Society and the Veterans Administration. Cancer Res. 1966. 26:2083–121.
8.Brett GZ. The value of lung cancer detection by six–monthly chest radiographs. Thorax. 1968. 23:414–20.

9.Wilde J. A 10 year follow-up of semi-annual screening for early detection of lung cancer in the Erfurt County, GDR. Eur Respir J. 1989. 2:656–62.
10.Frost JK., Ball WC Jr., Levin ML., Tockman MS., Baker RR., Carter D, et al. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Johns Hopkins study. Am Rev Respir Dis. 1984. 130:549–54.
11.Flehinger BJ., Melamed MR., Zaman MB., Heelan RT., Perchick WB., Martini N. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Memorial Sloan-Kettering study. Am Rev Respir Dis. 1984. 130:555–60.
12.Fontana RS., Sanderson DR., Taylor WF., Woolner LB., Miller WE., Muhm JR, et al. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Mayo Clinic study. Am Rev Respir Dis. 1984. 130:561–5.
13.Kubik A., Polak J. Lung cancer detection. Results of a randomized prospective study in Czechoslovakia. Cancer. 1986. 57:2427–37.
14.Fong KM., Sekido Y., Minna JD. Molecular pathogenesis of lung cancer. J Thorac Cardiovasc Surg. 1999. 118:1136–52.

15.Jones PA., Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999. 21:163–7. Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9: 359-67.
16.Wajed SA., Laird PW., DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001. 234:10–20.

17.Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999. 9:359–67.

18.Kamb A., Gruis NA., Weaver-Feldhaus J., Liu Q., Harshman K., Tavtigian SV, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994. 264:436–40.

19.Otterson GA., Kratzke RA., Coxon A., Kim YW., Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene. 1994. 9:3375–8.
20.Mao L., Hruban RH., Boyle JO., Tockman M., Sid-ransky D. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res. 1994. 54:1634–7.
21.Belinsky SA., Nikula KJ., Palmisano WA., Michels R., Saccomanno G., Gabrielson E, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998. 95:11891–6.

22.Esteller M., Sanchez–Cespedes M., Rosell R., Sidr-ansky D., Baylin SB., Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999. 59:67–70.
23.Ahrendt SA., Chow JT., Xu LH., Yang SC., Eisenberger CF., Esteller M, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999. 91:332–9.

24.Palmisano WA., Divine KK., Saccomanno G., Gilliland FD., Baylin SB., Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000. 60:5954–8.
25.Kersting M., Friedl C., Kraus A., Behn M., Pankow W., Schuermann M. Differential frequencies of p16(INK4-a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol. 2000. 18:3221–9.

26.Belinsky SA., Palmisano WA., Gilliland FD., Crooks LA., Divine KK., Winters SA, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002. 62:2370–7.
27.Destro A., Bianchi P., Alloisio M., Laghi L., Di Gioia S., Malesci A, et al. K-ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer. 2004. 44:23 -32.

28.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997. 111:1710–7.

29.Kunkel LM., Smith KD., Boyer SH., Borgaonkar DS., Wachtel SS., Miller OJ, et al. Analysis of human Y–chromosome–specific reiterated DNA in chromosome variants. Proc Natl Acad Sci U S A. 1977. 74:1245–9.

30.Herman JG., Graff JR., Myohanen S., Nelkin BD., Baylin SB. Methylation–specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996. 93:9821–6.

31.Mulshine JL., Treston AM., Scott FM., Avis I., Boland C., Phelps R, et al. Lung cancer: rational strategies for early detection and intervention. Oncology. 1991. 5:2. 5–32.
32.Wistuba II., Behrens C., Milchgrub S., Bryant D., Hung J., Minna JD, et al. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999. 18:643–50.

33.Geradts J., Fong KM., Zimmerman PV., Maynard R., Minna JD. Correlation of abnormal RB, p16ink4a, and p53 expression with 3p loss of heterozygosity, other genetic abnormalities, and clinical features in 103 primary non-small cell lung cancers. Clin Cancer Res. 1999 Apr. 5(4):791–800.
34.Kratzke RA., Greatens TM., Rubins JB., Maddaus MA., Niewoehner DE., Niehans GA, et al. Rb and p16INK4a expression in resected non–small cell lung tumors. Cancer Res. 1996 Aug 1. 56(15):3415–20.
36.Szyf M The DNA methylation machinery as a target for anticancer therapy. Pharmacol Ther. 1996. 70:1–37.
37.Antequera F., Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993. 90:11995–9.

39.Baylin SB. Tying it all together: epigenetics, genetics, cell cycle, and cancer. Science. 1997. 277:1948–9.

41.Lee JT., Jaenisch R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr Opin Genet Dev. 1997. 7:274–80.

44.Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987. 99:3. 71–82.

45.Lei H., Oh SP., Okano M., Juttermann R., Goss KA., Jaenisch R, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996. 122:3195–205.

46.Li E., Bestor TH., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992. 69:915–26.

48.Lee YW., Klein CB., Kargacin B., Salnikow K., Kitahara J., Dowjat K, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol. 1995. 15:2547–57.

49.Swafford DS., Middleton SK., Palmisano WA., Nikula KJ., Tesfaigzi J., Baylin SB, et al. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol. 1997. 17:1366–74.

50.Johnson TB., Coghill RD. The discovery of 5–methyl cytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J Am Chem Soc. 1925. 47:2838–44.
51.Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleotides by paper chromatography. J Biol chem. 1948. 175:315–32.
52.Srivastava R., Gopinathan KP., Ramakrishnan T. Deoxyribonucleic acid methylation in mycobacteria. J Bacteriol. 1981. 148:716–9.

53.Hemavathy KC., Nagaraja V. DNA methylation in mycobacteria: absence of methylation at GATC (Dam) and CCA/TGG (Dcm) sequences. FEMS Immunol Med Microbiol. 1995. 11:291–6.

54.Landis SH., Murray T., Bolden S., Wingo PA. Cancer statistics, 1998. CA Cancer J Clin. 1998. 48:6–29.

55.Ahuja N., Li Q., Mohan AL., Baylin SB., Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998. 58:5489–94.
56.Issa JP., Ottaviano YL., Celano P., Hamilton SR., Davidson NE., Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994. 7:536–40.

57.Melamed MR., Flehinger BJ., Zaman MB., Heelan RT., Perchick WA., Martini N. Screening for early lung cancer. Results of the Memorial Sloan-Kettering study in New York. Chest. 1984. 86:44–53.
58.Screening (lung cancer). Chest. 1986. 89:324S–326S.
Go to : 
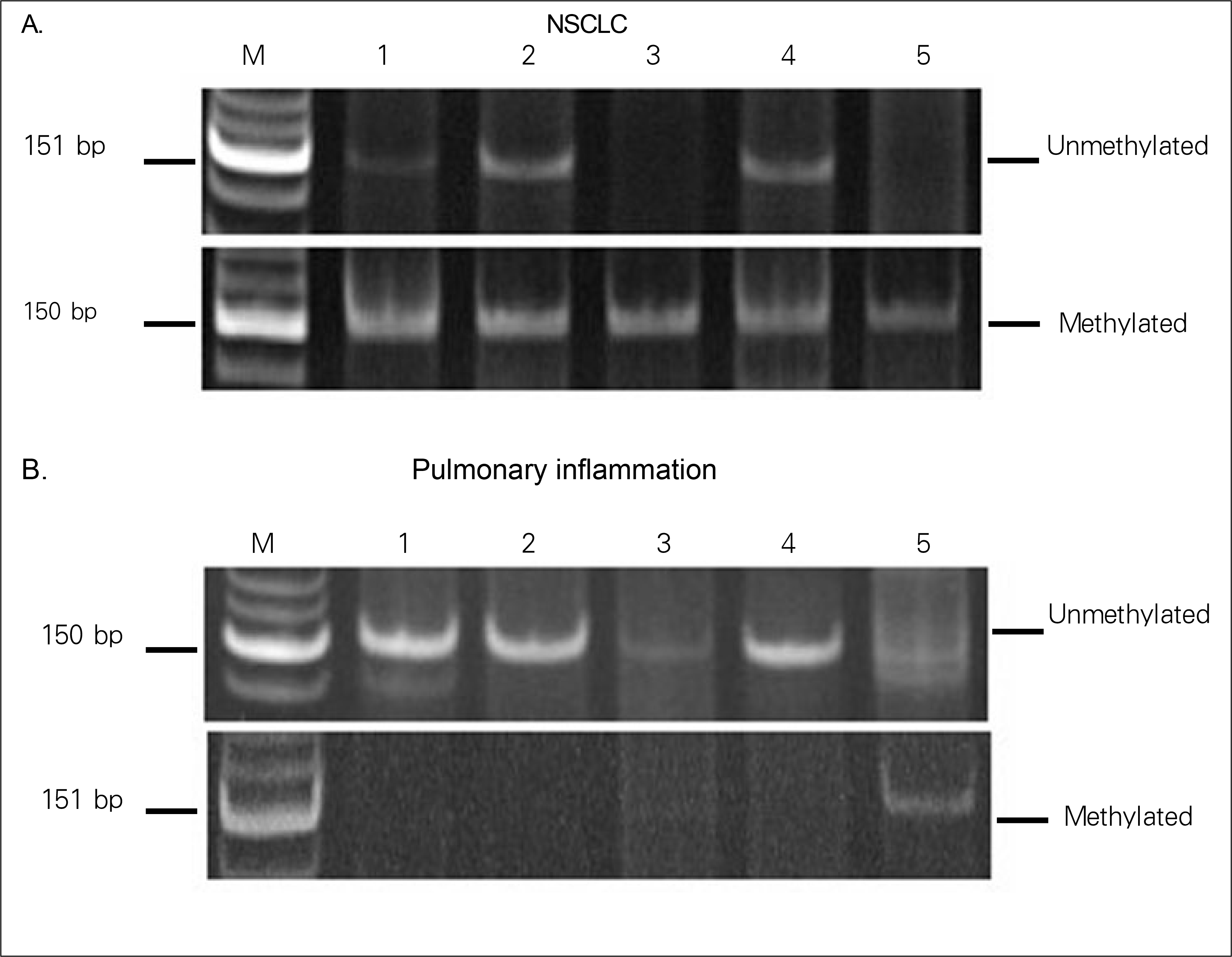 | Figure 1.Analysis of p16INK4a promoter hypermethylation in blood from NSCLC and pulmonary inflammation. Bisulfite modified DNA was amplified by primers specific to unmethylated or methylated alleles of p16INK4a. The prescence of visible PCR product in each lanes marked unmethylated or methylated. |
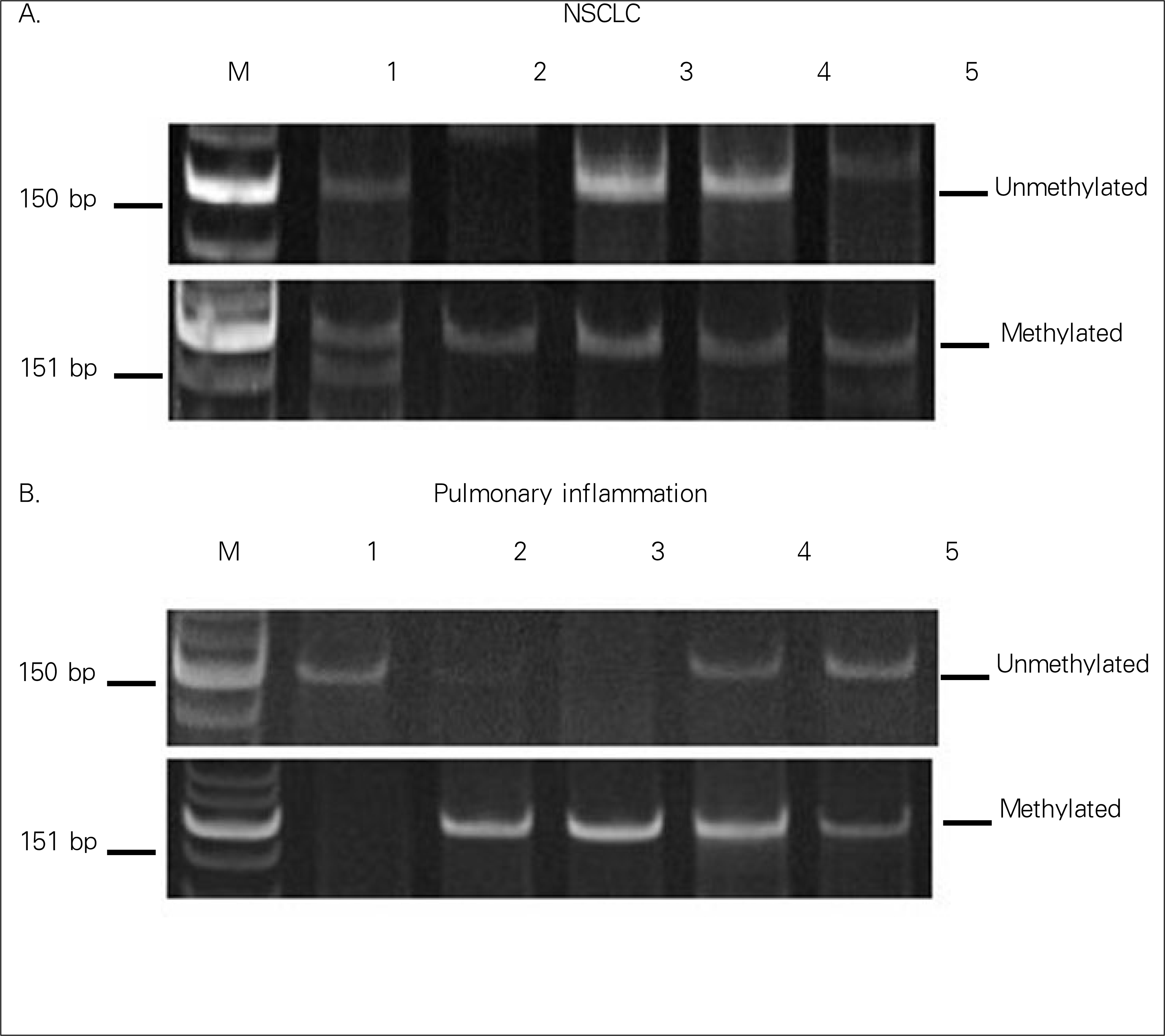 | Figure 2.Analysis of p16INK4a promoter hypermethylation in sputum from NSCLC and pulmonary inflammation. |
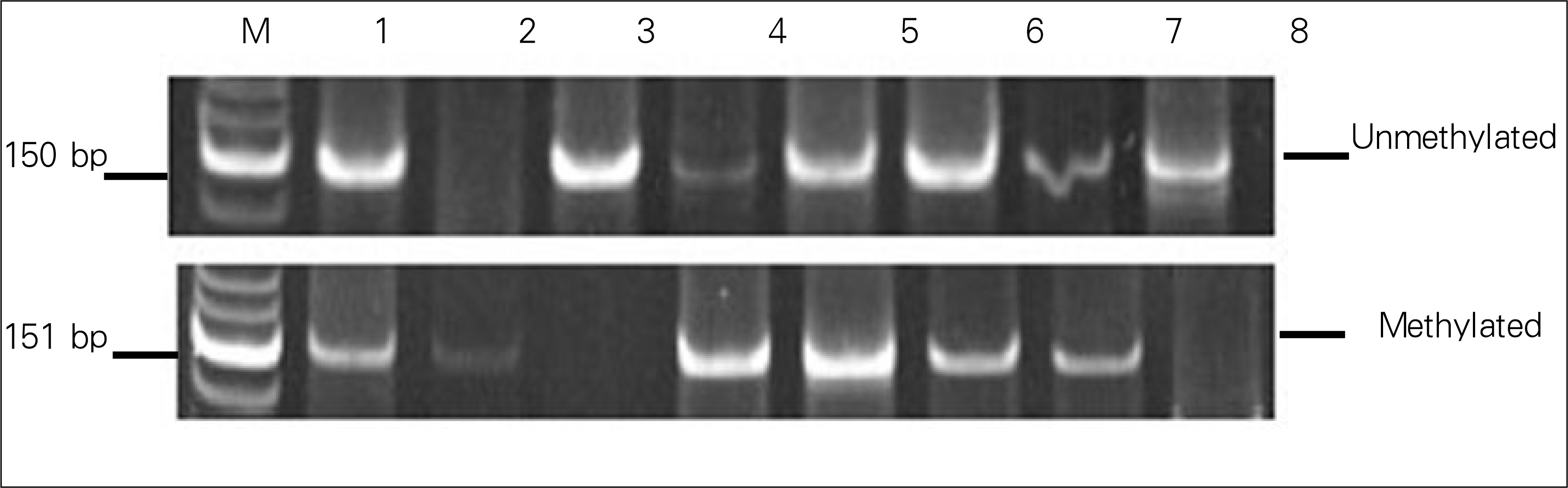 | Figure 3.Analysis of p16INK4a promoter hypermethylation in tissue from NSCLC. Methylation was observed in six of eight cases. |
Table 1.
Clinical characteristics of patients∗
| Characteristic | NSCLC† | Pulmonary inflammation | P value |
|---|---|---|---|
| No. of cases | 18 | 5 | |
| Age (yr) | 68±8 | 65±10 | 0.549 |
| Sex (no. of cases) | |||
| Male | 16 | 4 | 0.539 |
| Female | 2 | 1 | 0.539 |
| Smoking history | |||
| Pack-years | 39±25 | 24±25 | 0.329 |
| Current (no. of cases) | 12 | 1 | |
| Former | 4 | 2 | |
| Years since quitting | 10±13 | 10±13 | |
| Never | 2 | 2 | 4 |
Table 2.
p16INK4a hypermethylation in NSCLC∗ and pulmonary inflammation
| Sample (no. of cases) | NSCLC | Pulmonary inflmmation | P value |
|---|---|---|---|
| Blood | 16/18 (88.9%) | 1/5 (20.0%) | 0.008 |
| Sputum | 10/12 (83.3%) | 4/5 (80.0%) | 1.00 |
| Tissue | 6/8 (75.0%) | ‐ | ‐ |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download