Abstract
Clusterin (CLU) is a multifunctional glycoprotein that has secretory and nuclear isoforms. The two isoforms are known to play opposite roles in cell survival/death. In this review, we summarize recent progress on the pro-apoptotic function of nuclear CLU in vitro and in vivo and discuss previous reports on the role of CLU in brain damage and neurodegeneration.
Clusterin (CLU) was found initially as a cell-aggregating factor in ram rete testis fluid [1]. Two decades ago, CLU, a glycoprotein also known as apolipoprotein J, testosterone-repressed prostate message-2, and sulfate glycoprotein-2, was first cloned and characterized as a 427 amino acid polypeptide, which is post-translationally cleaved between residues 205 and 206 [2, 3]. The CLU transcript is expressed relatively highly in the brain, ovary, testis, and liver, and less abundantly in the heart, spleen, lung, and breast [3]. CLU was soon suggested as a marker of cell death, as it is upregulated in many cell types following cytotoxic stimulation [4-7]. However, accumulating results have revealed that CLU is a much more complicated protein than initially thought. In this review, we explore the pro-apoptotic role of nuclear CLU (nCLU) in vitro and in vivo in the context of brain damage and neurodegeneration.
Without glycosylation, human CLU is a protein of approximately 50 kDa and detected as a ~60 kDa glycosylated precursor secretory CLU (psCLU). It is cleaved to α and β chains of ~40 kDa and further glycosylated to form mature disulfide-linked heterodimeric secretory CLU (sCLU, 75-80 kDa). The nCLU transcript lacks the endoplasmic reticulum (ER)-targeting sequences at exon 2 and utilizes the second ATG site at exon 3 (Fig. 1A). The product is detected as the ~49 kDa nonglycosylated precursor nCLU in the cytosol and as a ~55 kDa glycosylated protein (nCLU) in the nucleus [8]. The CLU protein has two coiled coil domains responsible for interacting with other proteins, e.g., Ku70, which binds to CLU upon DNA damage [9] and two nuclear localization signals [7].
CLU plays controversial roles in apoptosis by producing two alternatively spliced isoforms in various cell types [10]. The pro-apoptotic CLU appears to be nCLU [7], and the sCLU and intracellular CLU are thought to be anti-apoptotic [11, 12]. The dichotomous roles of CLU in cell death were obvious from early studies using transgenic and gene-targeted mice, in which overexpression and deletion of the genes both caused reduced brain damage following hypoxia in vivo [13, 14]. Furthermore, previous reports showed that healthy human prostate cells expressed nCLU exclusively, but that human prostate cancer cells lost nCLU expression while increasingly expressing sCLU upon metastasis [12] and that deficiency of Clu enhances prostate cancer metastasis in prostate cancerprone transgenic adenocarcinoma of mouse prostate mice [15]. CLU translocation from the nucleus to the cytoplasm is related directly to colon tumorigenesis [16]. Considering these results, the expression and function of CLU in vivo must be finely tuned at the transcriptional, translational, and posttranslational levels.
As mentioned above, CLU is a bifunctional protein in terms of cell death and survival; sCLU or intracellular CLU inhibits apoptosis by interacting with Ku70 and Bax [17], whereas nCLU induces cell cycle arrest and cell death by inhibiting nuclear factor-κB-dependent Bcl-XL expression [18, 19]. In contrast to a previous study, in which intracellular CLU inhibited apoptosis by interacting with activated Bax [11], our group recently reported that nCLU sequestered Bcl-XL via a putative Bcl-2 homology 3 (BH3) domain, which was demonstrated in cells transfected with CLU gene deletion mutants [20] and by nuclear magnetic resonance spectroscopy [21]. Subsequently, Bax is released from Bcl-XL, promoting apoptosis accompanied by caspase-3 activation and cytochrome c release, implying that the role of nCLU is similar to derepressor/sensitizer BH3-only proteins [20].
BH3-only proteins are pro-apoptotic proteins with a single BH3 domain, unlike other Bcl-2 proteins that have multiple BH domains. Although the precise mechanism of the function of BH3-only proteins remains elusive, BH3-only proteins appear to interact with anti-apoptotic Bcl-2 proteins, e.g., Bcl-2 and Bcl-XL, and to activate effector Bcl-2 proteins, such as Bax and Bak, directly as an activator or indirectly as a derepressor/sensitizer, leading to cytochrome c release from mitochondria and cell death [22-27]. Overexpression of BH3-only proteins promotes apoptosis in many cell types but requires either Bax or Bak as an effector [28]. There are many BH3-only proteins, such as Bid, Bim, Bad, Bmf, Bik/Blk, Hrk, NOXA, and PUMA [29, 30]. Notably, BH3-only proteins do not usually have any other known functional domains rather than the BH3 domain [31]. In human nCLU, a putative BH3 motif was found in a C-terminal coiled coil (CC2) domain (Fig. 1A) [20, 21]. The sequence and tertiary structure of the putative BH3 motif were comparable to those of previously known BH3-only proteins (Fig. 1B). In particular, the hydrophobic L323 residue (in red Fig. 1B) and uncharged N328, which are highly conserved in BH3-only proteins, are indispensable for binding to Bcl-XL [20, 21]. Chemical shift perturbation data suggest that the nCLU BH3 domain could bind Bcl-2 as well [21], but that the actual interaction with Bcl-2 appears to be weaker than that of Bcl-XL [20].
We have also found that only nCLU, not sCLU, interacts with Bcl-XL [32]. Cleavage and heterodimerization to form sCLU could change protein folding and conformation, which might explain the differential roles of sCLU and nCLU in binding to Bax and Bcl-XL, respectively, and eventual cell survival/death. A recent report showed that the binding motif in psCLU was localized in the disulfide constraint region and was essential to bind IκB-α, but neither the α- nor the β-chain of sCLU had any activity [33]. This is another example to understand the importance of CLU protein conformation and tertiary structure. In addition, post-translational modifications could further regulate the differential roles of CLU. It is known that nCLU, located in the nucleus, is not glycosylated [34], whereas mature sCLU is heavily glycosylated [35].
CLU is expressed during fetal and postnatal central nervous system (CNS) development in mice [36]. CLU expression is upregulated in rats following traumatic brain injury [37], in seizure [38, 39], and in some neurodegenerative diseases, such as Parkinson's disease (PD) and Alzheimer's disease (AD) [40, 41]. The role of CLU in brain cell death is contradictory, as both gene-deficiency and overexpression of CLU inhibit brain damage in mice [13, 14]. It should be noted that the experimental conditions were different, as neonatal hypoxia/ischemia and permanent ischemia by middle cerebral artery occlusion in adult mice were used, respectively. CLU increases post-ischemic cell death in hippocampal slice cultures [42], implying a pro-apoptotic role for CLU.
We have discovered that nCLU plays a pro-apoptotic role in ethanol-induced cell death in the developing brain of infant rats [32]. We found that CLU increased in the cerebral cortex following acute ethanol treatment. CLU is broadly upregulated in the cerebral cortex, such as in Layers II, IV, and VI, and in the retrosplenial granular (RSG) cortex and the amygdala at 24 hour post-ethanol treatment. This CLU upregulation is related to increased cell death, which was assessed by activated caspase-3 levels and TUNEL staining. The upregulated CLU is a nuclear isoform that is translocated to the nucleus upon ethanol exposure and interacts with Bcl-XL, mediating apoptosis.
CNS dysfunction is one of the characteristics of fetal alcohol spectrum disorder (FASD), accompanied by abnormal brain structure/function, reduced white and grey matter volumes, and reduced total brain volume [43, 44]. It should be mentioned that our experimental model was heavy binge drinking, rather than chronic alcoholism. CLU expression was upregulated in the dispersed areas of the cerebral cortex, such as Layers II, IV, and VI, and in the amygdala upon acute ethanol exposure [32]. The amygdaloid complex is composed of more than ten nuclei, and one of the major functions of the amygdala in rats and humans is enhanced memory formation for emotional stimuli [45]. Upregulated CLU expression was found in the RSG cortex as well. The RSG cortex is an important link between hippocampal formation and other limbic areas of the brain and is equivalent to the cingulated gyrus in humans [46, 47]. It is also part of "Papez circuit," which was proposed by Papez as an emotion system [48]. The prefrontal cortex is involved in higher brain functions, including cognitive functions and behavioral control [49]. Furthermore, CLU upregulation is accompanied by upregulation of activated caspase 3 and TUNEL positivity in the brain following ethanol exposure, implying a role for CLU in brain cell death [32].
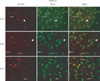
Additionally, we have found CLU upregulation and an interaction between CLU and Bcl-XL in the thalamus of the developing brain in infant rats upon ethanol exposure. As seen Fig. 2, CLU level increased post-ethanol exposure (left column), and CLU and Bcl-XL were co-localized (right column). Recent studies have reported that ethanol increases activated caspase-3 and subsequent neuronal cell death [50], which may be a result of ER stress induced by ethanol [51]. Nevertheless, the effect of ethanol on the thalamus is less understood. Our results suggest that CLU upregulation and interaction with Bcl-XL may mediate thalamic cell death by ethanol. The thalamus is located deep inside the forebrain and is often referred to as the gateway to the cerebral cortex, as thalamic neurons send axons to the cortex via the internal capsule [48]. In general, the internal capsule axons carry information to the cortex about the contralateral side of the body. It would be interesting to know the potential effect of ethanol on CLU expression and the interaction with other molecules and mechanisms thereof in the thalamus to understand the cause of behavioral deficits of FASD. Collectively, brain cell death mediated by nCLU in those areas might result in behavioral deficits and CNS abnormalities manifested in FASD. Furthermore, it is possible that CLU may be upregulated in other brain areas or cell types as well, such as the cerebellum, hippocampus, or microglial cells, which are known ethanol targets [52, 53]. Changes in CLU expression during chronic alcohol consumption have not been reported.
Not nCLU, but sCLU, has been suggested as a biomarker of neurodegenerative diseases, i.e., AD and PD, in the plasma or cerebrospinal fluid, respectively [40, 41]. In particular, CLU variants have been associated with AD in genome-wide association studies [54, 55], and plasma CLU levels reflect AD severity [40]. Therefore, CLU has been suggested as an AD risk gene. Intriguingly, sCLU, a pro-survival isoform, appears to fail to prevent the progression of neurodegeneration in AD, although it is upregulated in parallel to disease progression. In this context, sCLU might play a role as a chaperone in a similar manner to small heat shock proteins [56]. Expression and any contribution of nCLU need to be elucidated in neurodegenerative diseases as well. As nCLU plays a proapoptotic role, it is likely that nCLU expression could worsen neurodegeneration.
In conclusion, we have discovered the mechanism of the nCLU pro-apoptotic function in vitro and in vivo, which shows that nCLU interacts with Bcl-XL though a putative BH3 domain. Notably, all BH3-only proteins interact with and regulate the core Bcl-2 family proteins to promote apoptosis, and multiple BH3-only proteins are usually involved in cell death in one cell type [30]. Thus, it would be interesting to know whether nCLU and other BH3-only proteins are cooperative in the cell death process.
sCLU upregulation in AD is already suggested as a useful diagnostic and prognostic biomarker. nCLU expression and upregulation could be used as a biomarker of cell death and brain damage as well. More importantly, cell death could be enhanced or reduced by regulating nCLU expression. For example, inhibiting nCLU may reduce brain damage by seizure or ethanol and upregulating nCLU may reduce tumors. However, CLU has two isoforms and multiple binding proteins besides Bcl-XL. Although previous results imply that regulating CLU isoforms could change cell fate, the biological roles of CLU and the mechanism of cell death/survival regulated by CLU in vivo are thought to be more complex, depending on many factors, such as the expression/induction of each isoform, subcellular location of the protein, various stimuli, and the microenvironment. Thus, there is an urgent need to develop isoform-specific antibodies and siRNAs for further clarification of the biological roles of CLU in various cell types and conditions.
Figures and Tables
Fig. 1
Structure of nuclear clusterin (nCLU). (A) Schematic view of the CLU protein. The human CLU gene is located on chromosome 8 and consists of nine exons. The secretory CLU transcript starts with the first ATG (amino acid [a.a.] residue 1) and that of nCLU starts with the second ATG (a.a. 34). The CLU protein has two coiled coil (CC) domains and the putative Bcl-2 homology 3 (BH3) motif (a.a. 316-336) is in CC2. LP, leader peptide. (B) Sequence homolog y of BH3 motifs among BH3-only proteins. The core BH3 sequences are underlined, and the highly conserved arginine (L) is in red. The BH3 motif is responsible for binding the anti-apoptotic Bcl-2 family, such as Bcl-2 and Bcl-XL.

Fig. 2
Colocalization of clusterin (CLU) and Bcl-XL in the thalamus of the developing brain following ethanol treatment. Seven-day-old rats were subcutaneously injected with 6 g/kg of ethanol. After the designated times, they were sacrificed and perfused. Immunohistochemistry revealed that CLU expression was upregulated (left column) and colocalized with Bcl-XL (arrows) in the thalamus. The middle column show Bcl-XL expression, and the right column show merged images. E, ethanol treatment (×400).
Acknowledgements
We appreciate the former and present members of Prof. W. S. Choi's group for their constructive input. Prof. W. S. Choi's research was funded by the MRC program of the National Research Foundation of Korea (R13-2005-012-01001-1).
References
1. Blaschuk O, Burdzy K, Fritz IB. Purification and characterization of a cell-aggregating factor (clusterin), the major glycoprotein in ram rete testis fluid. J Biol Chem. 1983. 258:7714–7720.
2. de Silva HV, Stuart WD, Park YB, Mao SJ, Gil CM, Wetterau JR, Busch SJ, Harmony JA. Purification and characterization of apolipoprotein J. J Biol Chem. 1990. 265:14292–14297.
3. de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J. Apolipoprotein J: structure and tissue distribution. Biochemistry. 1990. 29:5380–5389.
4. Danik M, Chabot JG, Mercier C, Benabid AL, Chauvin C, Quirion R, Suh M. Human gliomas and epileptic foci express high levels of a mRNA related to rat testicular sulfated glycoprotein 2, a purported marker of cell death. Proc Natl Acad Sci U S A. 1991. 88:8577–8581.
5. Léger JG, Montpetit ML, Tenniswood MP. Characterization and cloning of androgen-repressed mRNAs from rat ventral prostate. Biochem Biophys Res Commun. 1987. 147:196–203.
6. Tenniswood MP, Guenette RS, Lakins J, Mooibroek M, Wong P, Welsh JE. Active cell death in hormone-dependent tissues. Cancer Metastasis Rev. 1992. 11:197–220.
7. Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem. 2003. 278:11590–11600.
8. Trougakos IP, Djeu JY, Gonos ES, Boothman DA. Advances and challenges in basic and translational research on clusterin. Cancer Res. 2009. 69:403–406.
9. Yang CR, Leskov K, Hosley-Eberlein K, Criswell T, Pink JJ, Kinsella TJ, Boothman DA. Nuclear clusterin/XIP8, an x-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci U S A. 2000. 97:5907–5912.
10. Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, Reichrath J. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006. 13:12–19.
11. Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005. 7:909–915.
12. Moretti RM, Marelli MM, Mai S, Cariboni A, Scaltriti M, Bettuzzi S, Limonta P. Clusterin isoforms differentially affect growth and motility of prostate cells: possible implications in prostate tumorigenesis. Cancer Res. 2007. 67:10325–10333.
13. Han BH, DeMattos RB, Dugan LL, Kim-Han JS, Brendza RP, Fryer JD, Kierson M, Cirrito J, Quick K, Harmony JA, Aronow BJ, Holtzman DM. Clusterin contributes to caspase-3-independent brain injury following neonatal hypoxia-ischemia. Nat Med. 2001. 7:338–343.
14. Wehrli P, Charnay Y, Vallet P, Zhu G, Harmony J, Aronow B, Tschopp J, Bouras C, Viard-Leveugle I, French LE, Giannakopoulos P. Inhibition of post-ischemic brain injury by clusterin overexpression. Nat Med. 2001. 7:977–979.
15. Bettuzzi S, Davalli P, Davoli S, Chayka O, Rizzi F, Belloni L, Pellacani D, Fregni G, Astancolle S, Fassan M, Corti A, Baffa R, Sala A. Genetic inactivation of ApoJ/clusterin: effects on prostate tumourigenesis and metastatic spread. Oncogene. 2009. 28:4344–4352.
16. Pucci S, Bonanno E, Pichiorri F, Angeloni C, Spagnoli LG. Modulation of different clusterin isoforms in human colon tumorigenesis. Oncogene. 2004. 23:2298–2304.
17. Trougakos IP, Lourda M, Antonelou MH, Kletsas D, Gorgoulis VG, Papassideri IS, Zou Y, Margaritis LH, Boothman DA, Gonos ES. Intracellular clusterin inhibits mitochondrial apoptosis by suppressing p53-activating stress signals and stabilizing the cytosolic Ku70-Bax protein complex. Clin Cancer Res. 2009. 15:48–59.
18. Scaltriti M, Santamaria A, Paciucci R, Bettuzzi S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells1. Cancer Res. 2004. 64:6174–6182.
19. Takase O, Minto AW, Puri TS, Cunningham PN, Jacob A, Hayashi M, Quigg RJ. Inhibition of NF-kappaB-dependent Bcl-xL expression by clusterin promotes albumin-induced tubular cell apoptosis. Kidney Int. 2008. 73:567–577.
20. Kim N, Yoo JC, Han JY, Hwang EM, Kim YS, Jeong EY, Sun CH, Yi GS, Roh GS, Kim HJ, Kang SS, Cho GJ, Park JY, Choi WS. Human nuclear clusterin mediates apoptosis by interacting with Bcl-XL through C-terminal coiled coil domain. J Cell Physiol. 2011. 05. 12. [Epub]. http://dx.doi.org/10.1002/jcp.22836.
21. Lee DH, Ha JH, Kim Y, Bae KH, Park JY, Choi WS, Yoon HS, Park SG, Park BC, Yi GS, Chi SW. Interaction of a putative BH3 domain of clusterin with anti-apoptotic Bcl-2 family proteins as revealed by NMR spectroscopy. Biochem Biophys Res Commun. 2011. 408:541–547.
22. Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001. 8:705–711.
23. Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002. 111:331–342.
24. Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005. 17:525–535.
25. Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007. 315:856–859.
26. Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009. 36:487–499.
27. Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006. 8:1348–1358.
28. Zong WX, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev. 2001. 15:1481–1486.
29. Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010. 37:299–310.
30. Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008. 9:47–59.
31. Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, Hinds MG. Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol. 2008. 380:958–971.
32. Kim N, Han JY, Roh GS, Kim HJ, Kang SS, Cho GJ, Park JY, Choi WS. Nuclear clusterin is associated with neuronal apoptosis in the developing rat brain upon ethanol exposure. Alcohol Clin Exp Res. 2011. 07. 18. [Epub]. http://dx.doi.org/10.1111/j.1530-0277.2011.01588.x.
33. Essabbani A, Margottin-Goguet F, Chiocchia G. Identification of clusterin domain involved in NF-kappaB pathway regulation. J Biol Chem. 2010. 285:4273–4277.
34. O'Sullivan J, Whyte L, Drake J, Tenniswood M. Alterations in the post-translational modification and intracellular trafficking of clusterin in MCF-7 cells during apoptosis. Cell Death Differ. 2003. 10:914–927.
35. Burkey BF, deSilva HV, Harmony JA. Intracellular processing of apolipoprotein J precursor to the mature heterodimer. J Lipid Res. 1991. 32:1039–1048.
36. Charnay Y, Imhof A, Vallet PG, Hakkoum D, Lathuiliere A, Poku N, Aronow B, Kovari E, Bouras C, Giannakopoulos P. Clusterin expression during fetal and postnatal CNS development in mouse. Neuroscience. 2008. 155:714–724.
37. Iwata A, Browne KD, Chen XH, Yuguchi T, Smith DH. Traumatic brain injury induces biphasic upregulation of ApoE and ApoJ protein in rats. J Neurosci Res. 2005. 82:103–114.
38. Noh HS, Kim DW, Kang SS, Cho GJ, Choi WS. Ketogenic diet prevents clusterin accumulation induced by kainic acid in the hippocampus of male ICR mice. Brain Res. 2005. 1042:114–118.
39. Schreiber SS, Tocco G, Najm I, Baudry M. Seizure activity causes a rapid increase in sulfated glycoprotein-2 messenger RNA in the adult but not the neonatal rat brain. Neurosci Lett. 1993. 153:17–20.
40. Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. JAMA. 2011. 305:1322–1326.
41. Přikrylová Vranová H, Mareš J, Nevrlý M, Stejskal D, Zapletalová J, Hluštík P, Kaňovský P. CSF markers of neurodegeneration in Parkinson's disease. J Neural Transm. 2010. 117:1177–1181.
42. Hakkoum D, Imhof A, Vallet PG, Boze H, Moulin G, Charnay Y, Stoppini L, Aronow B, Bouras C, Giannakopoulos P. Clusterin increases post-ischemic damages in organotypic hippocampal slice cultures. J Neurochem. 2008. 106:1791–1803.
43. Roebuck TM, Mattson SN, Riley EP. A review of the neuroanatomical findings in children with fetal alcohol syndrome or prenatal exposure to alcohol. Alcohol Clin Exp Res. 1998. 22:339–344.
44. Lebel C, Rasmussen C, Wyper K, Walker L, Andrew G, Yager J, Beaulieu C. Brain diffusion abnormalities in children with fetal alcohol spectrum disorder. Alcohol Clin Exp Res. 2008. 32:1732–1740.
45. Cahill L, McGaugh JL. Mechanisms of emotional arousal and lasting declarative memory. Trends Neurosci. 1998. 21:294–299.
46. Wyss JM, Van Groen T. Connections between the retrosplenial cortex and the hippocampal formation in the rat: a review. Hippocampus. 1992. 2:1–11.
47. Van Groen T, Wyss JM. Connections of the retrosplenial granular b cortex in the rat. J Comp Neurol. 2003. 463:249–263.
48. Bear MF, Connors BW, Paradiso MA. Neuroscience: exploring the brain. 2007. Philadelphia: Lippincott, Williams & Wilkins.
49. Weinberger DR, Egan MF, Bertolino A, Callicott JH, Mattay VS, Lipska BK, Berman KF, Goldberg TE. Prefrontal neurons and the genetics of schizophrenia. Biol Psychiatry. 2001. 50:825–844.
50. Young C, Roth KA, Klocke BJ, West T, Holtzman DM, Labruyere J, Qin YQ, Dikranian K, Olney JW. Role of caspase-3 in ethanol-induced developmental neurodegeneration. Neurobiol Dis. 2005. 20:608–614.
51. Ke Z, Wang X, Liu Y, Fan Z, Chen G, Xu M, Bower KA, Frank JA, Li M, Fang S, Shi X, Luo J. Ethanol induces endoplasmic reticulum stress in the developing brain. Alcohol Clin Exp Res. 2011. 35:1574–1583.
52. Suk K. Microglial signal transduction as a target of alcohol action in the brain. Curr Neurovasc Res. 2007. 4:131–142.
53. Norman AL, Crocker N, Mattson SN, Riley EP. Neuroimaging and fetal alcohol spectrum disorders. Dev Disabil Res Rev. 2009. 15:209–217.
54. Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P. European Alzheimer's Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009. 41:1094–1099.
55. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009. 41:1088–1093.
56. Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer's disease. Brain Res Rev. 2009. 61:89–104.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download