Abstract
Purpose
Molecular testing in non-small cell lung cancer (NSCLC) aids in identifying oncogenic alterations. The aim of this study was to compare the rates of detection of oncogenic alterations and responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) according to EGFR mutation status as determined by peptide nucleic acid (PNA) clamping or direct sequencing (DS).
Materials and Methods
We performed a systematic literature search using MEDLINE, EMBASE, and the Cochrane Central Register. Data from included studies were pooled to yield summary sensitivity, specificity, positive and negative likelihood ratios, diagnostic odds ratio, and receiver operating characteristic curves. A meta-regression analysis was conducted to identify potential sources of heterogeneity between selected studies.
Results
We identified 10 studies comprising 924 patients. Oncogenic alterations were detected in 340 of 924 cases (36.8%) with PNA clamping and in 250 of 924 (27.1%) with DS. The pooled sensitivities of PNA clamping and DS were 0.93 [95% confidence interval (CI): 0.90−0.95] and 0.69 (95% CI: 0.64−0.73), respectively. According to meta-regression analysis, none of the covariates were found to be significant sources of heterogeneity. With respect to treatment responses to EGFR-TKIs, there was no significant difference therein between EGFR mutations detected by PNA clamping and DS (53.4% vs. 50.8%; risk ratio, 0.99; 95% CI 0.83−1.19; p=0.874).
Lung cancer is the leading cause of cancer-related death worldwide.1 Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for approximately 85% of all cases.1 Since the discovery of oncogenic alterations in NSCLC, targeted therapeutic agents have been developed, resulting in improved survival in patients with treatable oncogenic alterations, including epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase translocation.2 Development of sensitive and specific methods to detect treatable oncogenic alterations is therefore a meaningful endeavor,2 and a variety of methods for detecting oncogenic alterations in NSCLC have been developed.3
Direct sequencing (DS) is the traditional and standard method for detecting EGFR mutations, and remains the most useful validated method for testing.3 However, DS requires a complex procedure, including DNA extraction, PCR-based amplification, DNA sequencing, and sequence interpretation.3 In addition, the sensitivity of DS is low, as it can only detect mutant DNA that makes up at least 25% of the total DNA content.4 Thus, several other assays have been created to increase sensitivity. The peptide nucleic acid (PNA) clamping method was recently approved by the Korean Food and Drug Administration.5 PNAs are artificially synthesized polymers that can strongly bind to complementary DNA sequences.6 PNA probes suppress PCR amplification of wild-type sequences, allowing for greater amplification of mutant sequences.6 Indeed, PNA clamping can be used to detect mutant alleles, even when present at levels 100-fold lower than wild-type alleles, and is less technically complex than DS.7 However, the PNA clamping method has a weakness in that it can only detect mutations for which primers have been individually designed.6 Thus, PNA clamping is not useful for detecting novel mutations.6
Whether intratumoral heterogeneity in EGFR mutations is one of the mechanisms for resistance to EGFR-tyrosine kinase inhibitors (TKIs) remains controversial.891011 Since responses to EGFR-TKIs could be correlated with EGFR mutant content, molecular tests with high sensitivity for detecting EGFR mutations might help with discerning which patients would be unlikely to respond to EGFR-TKIs.
DS has low sensitivity and has not been directly compared with PNA clamping methods in a systematic manner with respect to predicting oncogenic alterations. Furthermore, it is not clear if the PNA clamping method is non-inferior to DS with respect to predicting patient responses to EGFR-TKIs based on mutation detection. Thus, the primary aim of this study was to investigate whether PNA clamping has a higher rate of detection of oncogenic alterations, compared to DS, in patients with NSCLC. We also assessed clinical responses to EGFR-TKIs according to EGFR mutation status identified using both of these detection methods.
This meta-analysis was performed in accordance with the recommendations on the conduct and reporting of systematic reviews and meta-analyses outlined by the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement.12 To identify relevant articles eligible for this meta-analysis, we conducted a comprehensive search of three electronic databases (MEDLINE, EMBASE, and the Cochrane Central Register) up to September 01, 2017 using the following search terms: PNA clamping, DS, next-generation sequencing, pyrosequencing, lung cancer, lung adenocarcinoma, and NSCLC. As this study was a systematic review of published articles, neither informed consent nor ethics approval was required. The references listed in relevant review articles were also searched manually.
In order to be included in our analysis, studies had to meet the following inclusion criteria: 1) directly compared PNA clamping and DS; 2) included patients with a diagnosis of NSCLC; and 3) provided sufficient data to calculate absolute numbers of true-positive, false-positive, false-negative, and true-negative results. Studies published as full-length articles or letters in peer-reviewed English language journals were eligible for inclusion. Review articles, case reports, commentaries, and studies reporting outcomes but without raw data were excluded.
J-U.S. and J.L. independently conducted an extraction of potentially relevant articles, and reviewed each study in accordance with predefined eligibility criteria, after which data were extracted. Any disagreements that arose during the process of study selection or data extraction were resolved by consensus. A predefined form was used to extract data from each study. The information extracted from selected studies consisted of patient demographics, study design, and objectives.
As recommended by the Cochrane Collaboration, we used the Quality Assessment of Diagnostic Accuracy Studies (QUADAS)-2 tool to evaluate the risk of bias in diagnostic test accuracy.13 The QUADAS-2 tool consists of the four following key domains: patient selection, index test, reference standard, and flow/timing of patient selection. Supplementary Fig. 1 (only online) summarizes the seven items selected to evaluate risk of bias and applicability. The possible answers to each item were “yes,” “no,” or “unclear,” corresponding to a low, high, and unclear risk of bias, respectively. If a study was judged as “low” on all domains relating to bias or applicability, then it was assigned an overall judgment of “low risk of bias” or “low concern regarding applicability.” If a study was judged “high” or “unclear” in one or more domains, then it was judged as either “at risk of bias” or having “concerns regarding applicability.” Discrepancies were resolved by discussion between the two authors (J-U.S. and J.L.).
The data are presented as mean values for continuous variables and as frequencies (%) for categorical variables. For diagnostic meta-analysis, we extracted the numbers of patients with true-positive, false-positive, false-negative, and true negative test results either directly or through a recalculation based on the reported measures of accuracy in combination with the prevalence and sample size of the included study. We calculated pooled sensitivity and specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the receiver-operating characteristic curve (AUC) as point estimates with 95% confidence intervals (CIs).14 We additionally constructed summary receiver-operating characteristic curves (SROCs). In order to compare clinical responses to EGFR-TKIs according to detection method, we calculated pooled risk ratios (RR) and 95% CI. Between-study statistical heterogeneity was assessed using I2 and the Cochrane Q test.15 Heterogeneity was assessed using I2 statistics on a scale of 0−100%. A fixed-effects model was used unless I2 was >50%, indicating a substantial level of between-study heterogeneity, in which case a random-effects model was used.15
If substantial heterogeneity was found, analysis via meta-regression was performed to explore potential sources of bias.16 Publication bias was evaluated by Deeks' funnel plot asymmetry test.17 A p-value<0.05 was considered statistically significant. Statistical analyses were performed with Meta-Disc software (version 1.4, Madrid, Spain), Stata statistical software (version 14.2, StataCorp LP, College Station, TX, USA), and Review Manager (version 5.3, Nordic Cochrane Centre, the Cochrane Collaboration, Copenhagen, Denmark).
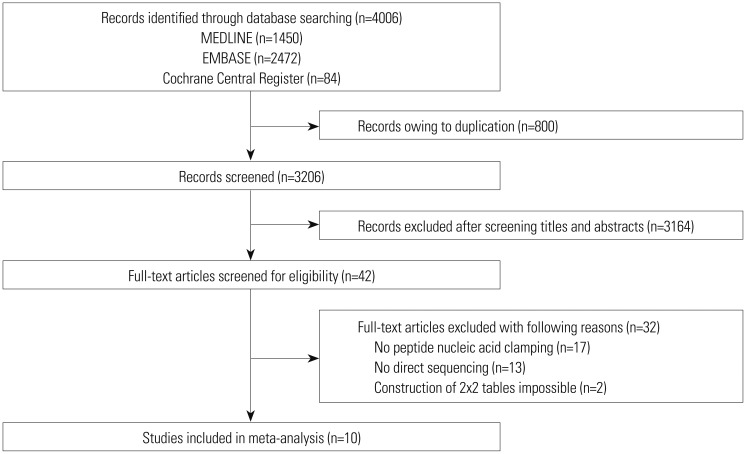
The literature search process is shown in Fig. 1. A total of 4006 published articles were initially identified (1450 articles from MEDLINE, 2472 articles from EMBASE, and 84 articles from the Cochrane library). After removing duplicate articles, we screened 3206 potentially eligible articles. After reviewing titles and abstracts, 3164 search records were removed, and the remaining 42 articles were further evaluated by reading the full text. Thirty-two articles were excluded for the reasons shown in Fig. 1. Finally, 10 studies were included in our final analysis.571819202122232425
Table 1 summarizes the features of the included studies. Ten studies involving 924 subjects met the defined inclusion criteria, and the numbers of patients in each trial ranged from 20 to 240. All studies were published between 2011 and 2016. Eight studies included results from NSCLC patients,57192021222325 and two studies included results solely from lung adenocarcinoma patients.1824 The rates of lung adenocarcinoma in the studies ranged from 58% to 100%.571819202122232425 Eight studies were evaluated for EGFR mutations, and the detection rates of EGFR mutation ranged from 35% to 62%.57182021222425 Two studies reported on KRAS mutations, and the detection rate of KRAS mutation in those two studies was 16%.1923
The QUADAS-2 assessment results are summarized in Supplementary Figs. 1 and 2 (only online). Overall, the quality of the included studies was deemed satisfactory. However, the QUADAS-2 tool identified unclear blinding during interpretation of results and a lack of reporting interpretable results as potential sources of bias. In addition, reasons for withdrawal were not clearly explained in some studies, which could have also resulted in bias.
Oncogenic alterations were detected in 340 of 924 cases (36.8%) with PNA clamping and in 250 of 924 cases (27.1%) with DS. With respect to PNA clamping, the pooled sensitivity and specificity for oncogenic alteration detection were 0.93 (95% CI, 0.90 to 0.95; I2=64.6%, p=0.003) and 1.00 (95% CI, 0.99 to 1.00; I2=0%, p=1.000), respectively (Fig. 2A and B). The pooled PLR and NLR were 73.03 (95% CI, 30.50 to 174.85; I2=0%, p=0.885) and 0.08 (95% CI, 0.04 to 0.16; I2=68.9%, p=0.001), respectively. The DOR for PNA clamping was 1138.36 (95% CI, 396.94 to 3264.581; I2=0%, p=0.509). For DS, the pooled sensitivity and specificity for detecting oncogenic alterations were 0.69 (95% CI, 0.64 to 0.73; I2=75.7%, p<0.001) and 1.00 (95% CI, 0.99 to 1.00; I2=0%, p=1.000), respectively (Fig. 2C and D). The pooled PLR and NLR were 52.59 (95% CI, 21.84 to 126.65; I2=0%, p=0.856) and 0.34 (95% CI, 0.24 to 0.47; I2=76.5%, p<0.001), respectively. The DOR for DS was 184.66 (95% CI, 71.22 to 478.81; I2=0%, p=0.623).
The SROC curves for the PNA clamping and DS tools with respect to predicting oncogenic alterations are shown in Fig. 3. The AUC was 0.996 [standard error (SE)=0.005] for PNA clamping and 0.971 (SE=0.042) for DS. Lastly, although Deeks' funnel plots showed asymmetric, there was no statistically significance of potential publication bias (p=0.07 for PNA clamping and p=0.15 for DS) (Supplementary Fig. 3, only online).
We retrieved data on the PNA clamping and DS molecular tests for detecting EGFR mutations from eight trials.57182021222425
EGFR mutations were detected in 316 of 770 cases (41.0%) with PNA clamping and in 228 of 770 cases (29.6%) with DS. For PNA clamping, the pooled sensitivity and specificity for detecting EGFR mutations were 0.92 (95% CI, 0.89 to 0.95; I2=67.8%, p=0.003) and 1.00 (95% CI, 0.99 to 1.00; I2=0%, p=1.000), respectively (Supplementary Fig. 4A and B, only online). The pooled PLR and NLR were 70.70 (95% CI, 26.74 to 186.89; I2=0%, p=0.856) and 0.08 (95% CI, 0.04 to 0.16; I2=72.3%, p=0.001), respectively. The DOR was 1113.71 (95% CI, 359.34 to 3451.76; I2=0%, p=0.546). For DS, the pooled sensitivity and specificity of the detection of EGFR mutations were 0.67 (95% CI, 0.62 to 0.72; I2=75.8%, p<0.001) and 1.00 (95% CI, 0.99 to 1.00; I2=0%, p=1.000), respectively (Supplementary Fig. 4C and D, only online). The pooled PLR and NLR were 47.52 (95% CI, 17.84 to 126.62; I2=0%, p=0.820) and 0.36 (95% CI, 0.27 to 0.49; I2= 76.6%, p<0.001), respectively. The DOR was 145.69 (95% CI, 51.95 to 408.57; I2=0%, p=0.688). The SROC curves for predicting EGFR mutations in the two groups are shown in Supplementary Fig. 5 (only online). The AUCs were 0.993 [standard error (SE)=0.009] for PNA clamping and 0.951 (SE=0.072) for DS.
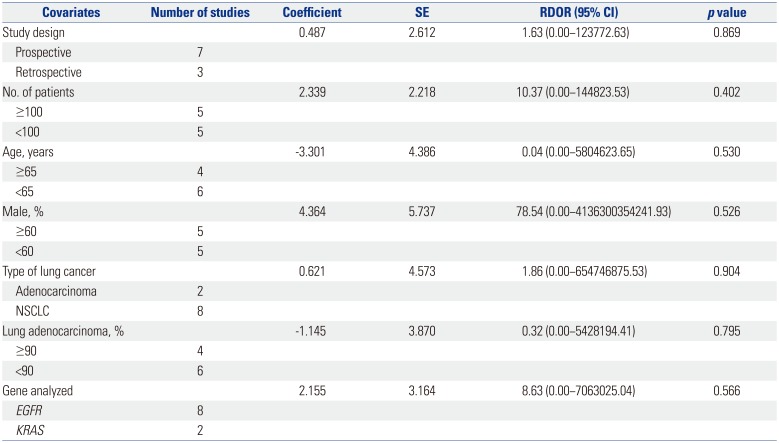
A substantial degree of heterogeneity was founded in the sensitivities and pooled LRs for both groups. Table 2 shows the results of meta-regression analyses for identifying potential sources of heterogeneity with respect to the diagnostic performance of PNA clamping for detecting oncogenic alterations in patients with NSCLC. Specifically, we investigated study design, sample size, age, proportion of males, type of lung cancer, rate of lung adenocarcinoma, and genes analyzed as probable sources of heterogeneity, and there were no significant factors identified.
We retrieved data on clinical responses to EGFR-TKIs from seven trials (Fig. 4).571820212225 Objective responses to EGFR-TKIs therapy for patients with EGFR mutations detected by PNA clamping were not significantly different from those of patients with EGFR mutations detected by DS (53.4% vs. 50.8%; RR, 0.99; 95% CI 0.83 to 1.19; I2=0%, p=0.874).
In the present study, we compared the effectiveness of DS and PNA clamping for predicting oncogenic alterations in patients with NSCLC using a systemic review and meta-analysis approach. Oncogenic alterations were detected in 340 of 924 cases (36.8%) with PNA clamping and in 250 of 924 cases (27.1%) with DS. We found that PNA clamping had a sensitivity of 93% for detecting oncogenic alterations, which was higher than the 69% sensitivity for DS. Subgroup analysis showed that PNA clamping had a sensitivity of 92% for detecting EGFR mutations, which was higher than the 67% sensitivity for DS. Although substantial heterogeneity was identified in the pooled estimates in both groups, potential sources of heterogeneity were not identified in a meta-regression analysis. In addition, there was no evidence of publication bias. Lastly, there was no significant difference in objective responses to EGFR-TKIs treatment between patients whose EGFR mutations were detected by PNA clamping or DS.
Molecular testing for oncogenic alterations has become an essential part of identifying patients eligible for treatment with targeted therapeutic agents. EGFR mutations are representative of treatable oncogenic alterations. Indeed, several randomized control trials have reported that NSCLC patients with EGFR mutations are more responsive to EGFR-TKIs and have a prolonged survival rate, compared to patients with non-mutated EGFR lung cancer.1 Thus, because EGFR mutation is a crucial prognostic biomarker for drug response, efficient detection of EGFR mutations is helpful for prolonging the survival of patients with NSCLC through targeted therapy.
DS can detect mutant DNA comprising ≥25% of the total DNA.26 However, the samples available for mutational analysis in lung cancer are frequently limited due to small tissue biopsy size or use of cytological specimens.18 Our results confirmed that the sensitivity of DS was low for detecting oncogenic alterations in NSCLC, and suggest the need for improved molecular testing methodologies. A number of sensitive detection methods have been developed based on the close relationship between treatable oncogenic alteration status and response to targeted agents. Among these methods, our pooled estimates demonstrated that PNA clamping was more sensitive than DS. In addition, PNA clamping is rapid and simple to conduct, and can detect mutant alleles comprising as little as approximately 1% of mutant alleles.6
Obtaining a sufficient amount of sample is critical for being able to perform both pathologic examination and mutation analysis. However, because many patients with NSCLC have advanced stage disease at the time of diagnosis, they frequently have inoperable disease and poor performance status. Thus, they often are not candidates for surgical treatment or invasive diagnostic procedures, which can often lead to a failure of obtaining a sufficient sample for analysis. Indeed, in some patients, the amount of diagnostic sample or cytological specimen is insufficient to permit additional molecular analysis after routine pathologic examination.19 Considering the frequency at which a very small amount of sample is obtained from NSCLC patients, PNA clamping has the potential to be very useful in clinical settings. Indeed, previous studies have revealed that the diagnostic performance of PNA clamping for detection of oncogenic alterations in NSCLC is favorable for cytology specimens, such as pleural effusions, as well as tumor tissue.719 However, to improve the rate of detection of oncogenic alterations, proper tissue handling, including timing of tissue sample acquisition, shorter fixation time, and DNA quality control, continue to be important.27
EGFR mutations can accumulate during tumor progression; thus, there is potential for heterogeneous EGFR status within tumors. In addition, multiple different EGFR mutations in a single tumor specimen have been reported; for this reason, heterogeneity of EGFR mutations has been suggested as one of the causes of resistance to EGFR TKIs.8910 Such intratumor heterogeneity of EGFR mutations could also partially explain why some patients with wildtype EGFR respond to EGFR-TKIs, whereas some patients with EGFR mutations fail to respond to EGFR-TKIs.18 A prospective study revealed that patients with heterogeneous EGFR mutation status do not respond well to EGFR-TKIs (gefitinib), and experience a significantly reduced time to progression and overall survival after gefitinib treatment, compared to patients with EGFR-mutant tumor cells.9 Because of the high sensitivity of PNA clamping, we were concerned that patients with a low burden of EGFR mutations detected by PNA clamping might also have a lower objective response to EGFR-TKIs than those detected by DS. However, according to pooled estimates, there was no significant difference in objective responses to EGFR-TKIs according to the method of mutation detection.
A recent prospective study reported that heterogeneity of EGFR mutations is extremely rare in lung adenocarcinoma, with the authors instead suggesting that reported findings of heterogeneity are better explain as “pseudoheterogeneity” attributable to mutant-allele specific imbalance and heterogeneously distributed EGFR amplification.11 In addition, because EGFR mutations can be detected in precancerous lesions, a large portion of invasive adenocarcinomas would contain EGFR mutations.11 The clinical implications of the difference between true heterogeneity and pseudoheterogeneity continue to be limited.18 Thus, further studies are needed to determine whether there are differences in other treatable oncogenic alterations detected by either DS or the PNA clamping method and if these are associated with differences in treatment responses to targeted therapy in NSCLC.
To the best of our knowledge, this is the first meta-analysis to compare PNA clamping and DS for predicting oncogenic alterations in NSCLC and to assess potential predictive biomarkers of response for EGFR-TKIs treatment. Our findings could be helpful for physicians implementing PNA clamping in clinical practice. However, there were some limitations to the present study that should be considered when interpreting our results. First, since our meta-analysis was based on a relatively small number of trials, our results should be interpreted with caution due to limited statistical power. Second, there was substantial heterogeneity among the included studies, although large heterogeneities are commonly seen in systematic reviews of studies on diagnostic test accuracy.28 Variations in baseline characteristics of the selected studies could have also potentially introduced heterogeneity. Third, 10 studies are too small to test for funnel plot asymmetry. Although there was no evidence of publication bias according to our analysis using the Deeks' test, the possibility could not be excluded. Considering of potential publication bias for small study effect, we additionally conducted the Begg's test and the Egger's test. For PNA clamping and DS, p-values of the two tests indicated that there also was no publication bias in the meta-analysis (0.592 and 0.691 for the Begg's test and 0.721 and 0.724 for the Egger's test, respectively).
In conclusion, PNA clamping was found to be able to detect oncogenic alterations in an additional 100 out of approximately 920 patients with NSCLC, compared to DS. We demonstrated that PNA clamping had higher sensitivity and accuracy than DS for predicting oncogenic alterations in NSCLC. With respect to objective responses to EGFR-TKIs therapy, there were no significant differences between patients whose EGFR mutations were detected by PNA clamping or DS. Our findings provide evidence that PNS clamping is more useful than DS among the current methods for molecular testing in NSCLC.
References
1. Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017; 377:849–861. PMID: 28854088.

2. Won JK, Keam B, Koh J, Cho HJ, Jeon YK, Kim TM, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol. 2015; 26:348–354. PMID: 25403583.

3. Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013; 137:828–860. PMID: 23551194.

4. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007; 13:4954–4955. PMID: 17785543.
5. Kim HS, Sung JS, Yang SJ, Kwon NJ, Jin L, Kim ST, et al. Predictive efficacy of low burden EGFR mutation detected by next-generation sequencing on response to EGFR tyrosine kinase inhibitors in non-small-cell lung carcinoma. PLoS One. 2013; 8:e81975. PMID: 24376508.

6. Lee KY, Kim HJ, Kim SJ, Yoo GH, Kim WD, Oh SY, et al. PNA-mediated PCR clamping for the detection of EGFR mutations in non-small cell lung cancer. Tuberc Respir Dis. 2010; 69:271–278.

7. Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ, et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2013; 81:207–212. PMID: 23726527.

8. Sakurada A, Lara-Guerra H, Liu N, Shepherd FA, Tsao MS. Tissue heterogeneity of EGFR mutation in lung adenocarcinoma. J Thorac Oncol. 2008; 3:527–529. PMID: 18449007.

9. Taniguchi K, Okami J, Kodama K, Higashiyama M, Kato K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008; 99:929–935. PMID: 18325048.

10. Yeh YC, Chou TY. Pulmonary adenocarcinoma with microcystic histology and intratumoral heterogeneity of EGFR gene polymorphism. Histopathology. 2010; 57:112–120. PMID: 20653783.

11. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011; 29:2972–2977. PMID: 21730270.

12. Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009; 151:264–269. PMID: 19622511.

13. Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003; 3:25. PMID: 14606960.

14. Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005; 58:982–990. PMID: 16168343.

15. Thompson SG. Why sources of heterogeneity in meta-analysis should be investigated. BMJ. 1994; 309:1351–1355. PMID: 7866085.
16. Lijmer JG, Mol BW, Heisterkamp S, Bonsel GJ, Prins MH, van der Meulen JH, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA. 1999; 282:1061–1066. PMID: 10493205.

17. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005; 58:882–893. PMID: 16085191.

18. Han HS, Lim SN, An JY, Lee KM, Choe KH, Lee KH, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012; 7:355–364. PMID: 22157369.

19. Kang JY, Park CK, Yeo CD, Lee HY, Rhee CK, Kim SJ, et al. Comparison of PNA clamping and direct sequencing for detecting KRAS mutations in matched tumour tissue, cell block, pleural effusion and serum from patients with malignant pleural effusion. Respirology. 2015; 20:138–146. PMID: 25302858.
20. Kim HJ, Kim WS, Shin KC, Lee GH, Kim MJ, Lee JE, et al. Comparative analysis of peptide nucleic acid (PNA)-mediated real-time PCR clamping and DNA direct sequencing for EGFR mutation detection. Tuberc Respir Dis. 2011; 70:21–27.

21. Kim HJ, Lee KY, Kim YC, Kim KS, Lee SY, Jang TW, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012; 75:321–325. PMID: 21930325.

22. Lee HJ, Xu X, Kim H, Jin Y, Sun P, Kim JE, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60. PMID: 23483646.
23. Lee B, Lee B, Han G, Kwon MJ, Han J, Choi YL. KRAS mutation detection in non-small cell lung cancer using a peptide nucleic acid-mediated polymerase chain reaction clamping method and comparative validation with next-generation sequencing. Korean J Pathol. 2014; 48:100–107. PMID: 24868222.
24. Min KW, Kim WS, Jang SJ, Choi YD, Chang S, Jung SH, et al. Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study. QJM. 2016; 109:167–173. PMID: 26031706.

25. Yoon SH, Choi YD, Oh IJ, Kim KS, Choi H, Chang J, et al. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non-small cell lung cancer. Cancer Res Treat. 2015; 47:661–669. PMID: 25715768.
26. Endo K, Konishi A, Sasaki H, Takada M, Tanaka H, Okumura M, et al. Epidermal growth factor receptor gene mutation in non-small cell lung cancer using highly sensitive and fast TaqMan PCR assay. Lung Cancer. 2005; 50:375–384. PMID: 16199108.

27. Pirker R, Herth FJ, Kerr KM, Filipits M, Taron M, Gandara D, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010; 5:1706–1713. PMID: 20871269.

SUPPLEMENTARY MATERIALS
Supplementary Fig. 1
Summary of risk of bias in the included studies. The green, yellow, and red symbols represent a low, unclear, and high risk of bias, respectively. The figure was generated using Review Manager version 5.3.
Supplementary Fig. 2
Risk of bias graph for the included studies. Each methodological quality item is presented as a percentage across all included studies. The figure was generated using Review Manager version 5.3.
Supplementary Fig. 3
Funnel plot for publication bias assessment of studies for peptide nucleic acid clamping (A) and direct sequencing (B) for detection of oncogenic alterations. ESS, effective sample size; DOR, diagnostic odds ratio.
Supplementary Fig. 4
Paired forest plots of sensitivity and specificity of peptide nucleic acid clamping (A and B) and direct sequencing (C and D) for detection of EGFR mutations, CI, confidence interval.
Supplementary Fig. 5
SROC curves for peptide nucleic acid clamping (A) and direct sequencing (B) for detection of EGFR mutations. SROC, summary receiver operating characteristic; AUC, area under the curve; SE, standard error.
Fig. 2
Paired forest plots of the sensitivity and specificity of peptide nucleic acid clamping (A and B) and direct sequencing (C and D) for the detection of oncogenic alterations. CI, confidence interval.
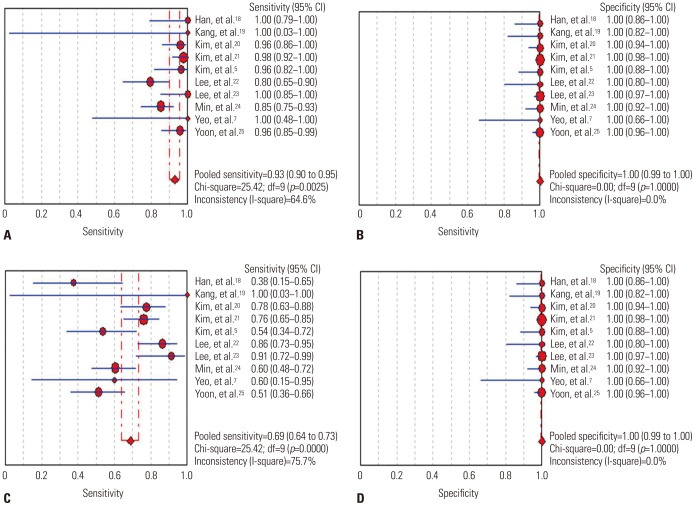
Fig. 3
SROC curves for peptide nucleic acid clamping (A) and direct sequencing (B) for the detection of oncogenic alterations. SROC, summary receiver operating characteristic; AUC, area under the curve; SE, standard error.
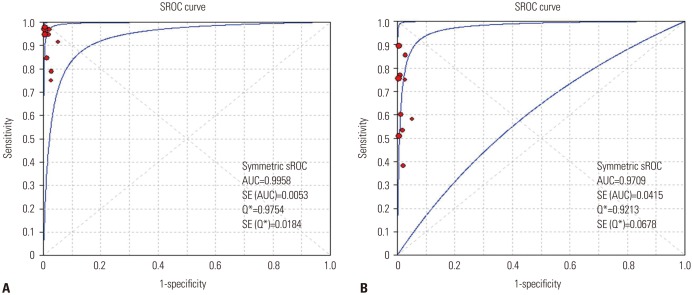
Fig. 4
Pooled results of the response to EGFR-tyrosine kinase inhibitors in EGFR activating mutation-positive non-small cell lung cancer detected by PNA clamping and DS. EGFR, epidermal growth factor receptor; PNA, peptide nucleic acid; DS, direct sequencing; RR, risk ratio; CI, confidence interval.
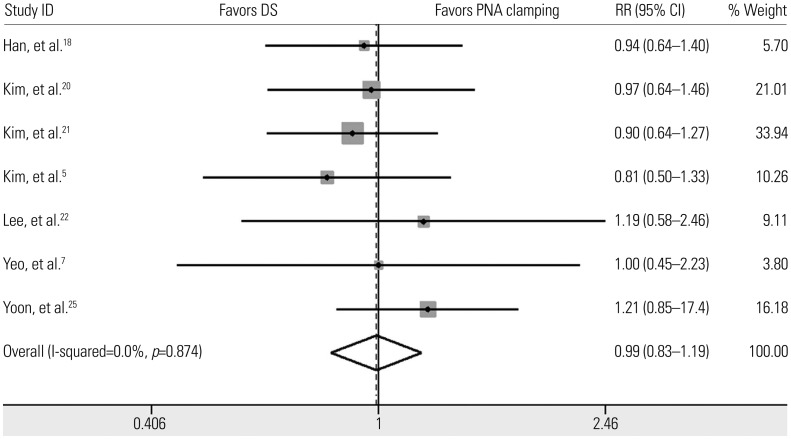
Table 1
Characteristics of the Studies Included in the Meta-Analysis
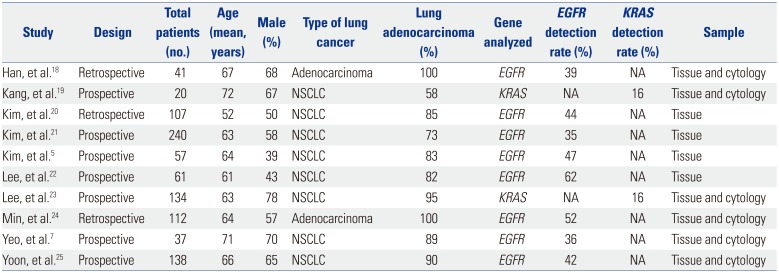
| Study | Design | Total patients (no.) | Age (mean, years) | Male (%) | Type of lung cancer | Lung adenocarcinoma (%) | Gene analyzed | EGFR detection rate (%) | KRAS detection rate (%) | Sample |
|---|---|---|---|---|---|---|---|---|---|---|
| Han, et al.18 | Retrospective | 41 | 67 | 68 | Adenocarcinoma | 100 | EGFR | 39 | NA | Tissue and cytology |
| Kang, et al.19 | Prospective | 20 | 72 | 67 | NSCLC | 58 | KRAS | NA | 16 | Tissue and cytology |
| Kim, et al.20 | Retrospective | 107 | 52 | 50 | NSCLC | 85 | EGFR | 44 | NA | Tissue |
| Kim, et al.21 | Prospective | 240 | 63 | 58 | NSCLC | 73 | EGFR | 35 | NA | Tissue |
| Kim, et al.5 | Prospective | 57 | 64 | 39 | NSCLC | 83 | EGFR | 47 | NA | Tissue |
| Lee, et al.22 | Prospective | 61 | 61 | 43 | NSCLC | 82 | EGFR | 62 | NA | Tissue |
| Lee, et al.23 | Prospective | 134 | 63 | 78 | NSCLC | 95 | KRAS | NA | 16 | Tissue and cytology |
| Min, et al.24 | Retrospective | 112 | 64 | 57 | Adenocarcinoma | 100 | EGFR | 52 | NA | Tissue and cytology |
| Yeo, et al.7 | Prospective | 37 | 71 | 70 | NSCLC | 89 | EGFR | 36 | NA | Tissue and cytology |
| Yoon, et al.25 | Prospective | 138 | 66 | 65 | NSCLC | 90 | EGFR | 42 | NA | Tissue and cytology |
Table 2
Meta-Regression Analysis to Identify Potential Sources of Heterogeneity




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download