I. Introduction
Tumors develop when the balance between oncogenes that induce tumors and tumor suppressor genes that repress them is disturbed. A normal cell transforms into a cancer cell when various genetic modifications take place; these lessen the control on cell cycles and lead to abnormal cell growth, which gives rise to tumors.
Previously, such control mechanisms were known to be affected by the change in DNA sequence and recombination, which cause a change in phenotype; nowadays, however, it is known that gene function may change without DNA sequence modifications, and that this change is passed on from parents to offspring. Epigenetics is a new field of study that investigates whether such change in gene function occurs without DNA sequence modifications and includes various modification processes such as DNA acetylation, methylation, phosphorylation, and ubiquitination. Well-known studies are those exploring the gene control mechanism concerning DNA methylation
1.
Numerous new findings are being made in the studies on tumors through epigenetics, including genetic variations in cells that express tumors. In a general genetic aspect, the mutation of DNA base is an important phenomenon; from an epigenetic viewpoint, however, the methylation process-which is the attachment of a methyl group to the base by a DNA methyltransferase (DNMT)-is significant
1,
2. This process takes place only when a cytosine (C) is followed by a guanine (G), which is called CpG when the two bases are situated next to each other in a human genome DNA sequence; when CpGs are densely located, such is called a CpG island. This CpG island is located near the promoter area that controls the transcription process of the DNA. In particular, cytosine tends to be methylated, with 3-4% of the total cytosine of the human genome being methylated. Although it differs according to the type of tumor, in the development of a malignant tumor, oncogenes are activated after hypomethylation, whereas tumor suppressor genes are inactivated with hypermethylation. Studies report that this change in methylation is associated with the development of tumor
1,
3.
One of the tumor suppressor genes, p16
INK4a (Inhibitor of cyclin-dependent kinase 4/6), encodes p16 proteins. The promoter area for the p16
INK4a gene includes the DNA sequence for the production of protein p16. The inactivation of this gene plays an important role in the development of a malignant tumor. The loss of p16 results from point mutation
4, loss of heterozygosity (LOH)
5, or hypermethylation
1-
5 of the promoter area. In this study, we investigated the presence of p16 protein and hypermethylation that inactivates the p16
INK4a gene in patients with head and neck squamous cell carcinoma in a retrospective design and compared it with the clinical findings of the tumor, recurrence, and survival rate. Specifically, we tried to determine whether epigenetic gene modifications have a direct effect on protein expression and decide whether the methylation of the tumor suppressor gene, p16, affects the prognosis of head and neck squamous cell carcinoma.
II. Materials and Methods
1. Materials
Tissue samples of 59 patients with head and neck squamous cell carcinoma and who had been diagnosed, operated, and subjected to post-operative evaluations at the Department of Oral and Maxillofacial Surgery, Pusan National University were used for the immunohistochemical tests of p16 protein and investigation of methylation of the p16INK4a gene. The tissue was fixed with formalin and embedded in paraffin and stored in the Department of Pathology, Pusan National University Hospital. There were 15 women and 44 men, with mean age of 61.8 years (range: 28-86 years). The tumor sample was taken from a biopsy done before the initiation of treatment. All diagnoses were made based on samples with H&E staining along with histopathological tests.
2. Clinical and histopathological tests
The age of the patient, tumor location and size (4 grades: less than 2 cm, less than 4 cm, more than 4 cm, and invasion into adjacent structures), existence of smoking habit (+ vs -), recurrence (+ vs -), level of differentiation (well, moderate, or poor), and pathological stage (stage I-stage IV: American Joint Committee on Cancer [AJCC]
6 6th edition) were investigated and grouped for comparison with the expression of p16 protein and methylation of the p16
INK4a gene. Survival rate was determined in all cases, with observation period of 0.5-86 months (mean: 26.8 months). The size of the tumor was judged and grouped based on visual inspection on the first visit and computed tomography (CT) and magnetic resonance imaging (MRI) images, with pathological staging and grouping done based on general clinical examinations along with visual inspection on the first visit, chest radiographs, laboratory findings, CT and MRI images, location and size of lymph, and occurrence of metastasis. The correlation between the p16
INK4a gene and smoking as a major carcinogenic factor of intraoral cancer was also investigated.
3. Immunohistochemical tests
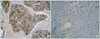
Immunohistochemical tests were done by gaining 4-µm thick sections of the paraffin-embedded tissues and attaching the tissue to a poly-L-lysine-coated slide and heating in an 80℃ oven for 15 minutes and then removing the paraffin by treating for 2 minutes for 4 times at room temperature with xylene and soaking in numerous stages of alcohol and rehydrating with distilled water. To recover antigenicity, the sample was soaked in 10% citrate butter and boiled in a microwave oven for 10 minutes, and then made to stand at room temperature for 20 minutes followed by washing with 45℃ buffered solution for 3 minutes thrice. Afterward, to remove intrinsic tissue peroxidase, the tissue was left in 3% H2O2 solution for 5 minutes. The sample was then washed with buffered solution thrice, and non-immunologic goat serum (Zymed Laboratory Inc., San Francisco, CA, USA) was applied to remove the linkage of nonspecific proteins. The sample was left overnight at 4℃ to react with p16 (PharMingen, San Diego, CA, USA) as the primary antibody. The secondary antibody- anti-mouse IgG human monoclonal antibody (NeoMarkers, Fremont, CA, USA)-was then applied for 10 minutes. The samples were washed with buffered solution for 3 minutes thrice and treated with a Streptavidin-biotin peroxidase kit (Zymed Laboratory, Inc., San Francisco, CA, USA) at 45℃ for 10 minutes to induce biotin-streptavidin specific linkage. The sample was then washed again, and color reaction was induced with 3-amino-9-ethylcarbazole for 10 minutes; counterstaining was done with hematoxylin before mounting and observing with a light microscope. The palatine tonsil tissue was used as positive control, and tissues that were not treated with the primary antibody were used as negative control.
Each case was scored according to the decided criteria
7-
9. For example, if the nucleus of the tumor cells was evenly stained throughout the entire tumor, p16 expression was considered normal. In contrast, the expression of p16 was deemed to have decreased in that tumor when the nuclei of the mixed non-tumor cells were stained and those of the tumor cells were only partially stained or the nuclei of the mixed non-tumor cells were stained but those of the stained tumor cells were missing.
4. DNA extraction from tissue samples
Genomic DNA was extracted with a standardized proteinase K digestion and the phenol/chloroform extraction method. The tissue was taken from tumor locations with distinct pathohistological characteristics of squamous cell carcinoma, and DNA was extracted from this sample for PCG assay.
5. Bisulphite modification and methylation-specific polymerase chain reaction (MSP)
Bisulphite modification was done with a DNA modification kit (Intergen, Purchase, NY, USA) by following the manufacturer's instructions. The modified DNA was used for MSP.
Polymerase chain reaction (PCR) was done in 35 cycles at 95℃ following the first denaturation (95℃ for 45 seconds, 60℃ for 45 seconds, and 72℃ for 60 seconds). The samples were then left at 72℃ for the final 4 minutes. Colo-205 cell line DNA, which shows hypermethylation of the p16
INK4a gene promoter area, was used as positive control according to the MSP method
9. Additionally, DNA extracted from normal skeletal muscles was used as negative control. Each PCR product (10 µL) was loaded on 10% polyacrylamide gel directly, stained with ethidium bromide, and then observed under ultraviolet lights directly.
6. Statistical analysis
Fisher's exact test was used to analyze the correlation between clinicopathological factors including age, gender, recurrence, and smoking habit according to p16 methylation and immunohistochemical expression of the p16 gene. The chi-square test was used for factors including tumor size, pathological stage, and stage of differentiation. Analysis involving survival rate was done with SAS version 9.13 (SAS Institute, Inc., NC, USA) using the Kaplan-Meier survival curve method
10 to investigate the survival rate according to p16 methylation and immunohistochemical expression of the p16 gene. The differences in survival rate between the methylation and non-methylation, and between immunohistochemical expression and immunohistochemical non-expression groups were analyzed with log-rank test. The results were considered statistically significant when
P-value<0.05.
IV. Discussion
The genetic information of a somatic cell is transferred to the next generation through a series of cell division. This cell division is repeated in cycles, and such replication period is called a cell cycle. This phenomenon consists of a train of regular biological events that follow an exact timetable, which include gene expression and regular changes in protein phosphorylation.
Some factors stimulate cell proliferation, whereas other factors suppress it during the cell growth control process. Growth factors and their receptors and constituents of the mitogen-activated protein kinase pathway stimulate cell growth and division, whereas repressors of the cyclin-cyclin-dependent kinase (CDK) complex, retinoblastoma protein (pRb), and proteins such as p53 suppress it. Most tumors are caused by somatic mutations of cell genes encoding protein products that stimulate or suppress cell growth.
The cell cycle is generally divided into cell division interphase (G1-S-G2) and mitosis (M); at 2 points during this cycle, an all-or-nothing decision is made as to whether a normal cell cycle will proceed or stop. First is the G1 checkpoint, which happens before moving on to S stage from G1 stage; the second point is the G2 checkpoint, which happens before entering the mitosis phase. Among these, pRb, p16, and cyclin D are known as key proteins that control cell growth in the G1 checkpoint
11,
12.
In particular, the p16
INK4a gene is located at 9p21 on the short hand of the ninth chromosome, stopping the process of the cell cycle and encoding a nuclear protein that represses cell growth by bonding with CDK4/6 to interrupt the bonding of cyclin and suppressing CDK activity
13-
16. When a functional p16 is absent, however, CDK4/6 bonds to cyclin D and phosphorylates pRb, resulting in the release of the transcription factor E2F that stimulates the process of the cell cycle. Many reports have shown such activation in malignant tumors, suggesting that the reduction in p16 expression is related to the progression of tumors and survival rate in many types of tumors
7,
17-
19. Moreover, the loss of the functional p16
INK4a gene is frequently seen in various tumors and malignancies, which probably results from LOH, point mutation, and methylation of the promoter area. From a general genetic viewpoint, the mutation of base substitution is an important phenomenon; in epigenetics, however, methylation of the DNA base by attachment of a methyl group has huge significance
1,
2.
Epigenetic modifications such as the methylation of DNA supposedly affect the expression or progression of various tumors. This occurs through the reversible attachment of a methyl group to the 5th carbon of a cytosine in the CpG island, serving as an important process in the development of a normal embryo. When the general hypomethylation of DNA and the abnormal hypermethylation of the tumor suppressor gene promoter area destabilize the chromosome and decrease transcription, however, a selectively advan-tageous environment for cells of a neoplasm is created
20.
Hypermethylation of the CpG island near the p16INK4a gene promoter area neutralizes the function of migratory genes such as a transposon that flows in from the exterior and acts as a defense mechanism by the loss of transcription. This is believed to occur by the substitution of methylated cytosine with thymine, which results in the gradual loss of function of migratory genes over time.
Three types of epigenetic information associated with the methylation of genes show high correlation: methylation of DNA, histone modification, and genomic imprinting
1,
2.
The methylation of DNA is carried out by DNMT. Methy-lation disturbs the recognition of transcription factors. Once DNA is methylated, "methylated protein" is induced from this region. These proteins attach to factors such as "histone deacetylase" and create a complex. The methylation of DNA suppresses the transcription process by histone modification or chromatin remodeling rather than DNMT activation. Thus, the three different mechanisms- DNA methylation, histone methylation or deacetylation, and chromatin remodeling- are closely related. Moreover, the methylation of DNA maintains gene silence. With demethylation by the removal of DNMT, gene reactivation is observed
1,
2.
When the chromosomes inherited from the paternal and maternal line all function, there are scores of gene group types that produce genetic defects. In such cases, genes of only one side should be expressed; this control to a proper state is called genomic imprinting. Genes controlled in such fashion are called imprinted genes. However, genetic abnor mality occurs when there is loss of imprinting and the imprinted gene appears to be methylated
1.
Various previous studies have reported the effect of p16
INK4a gene methylation on carcinogenesis. Liu et al.
21 measured the level of p16 gene methylation of normal mucosa, leukoplakia, which is a precancerous lesion, and each stage of its cancerous change to oral squamous cell carcinoma and reported that the portion of p16 gene methylation increased with the grade of malignancy of the tissue. Furthermore, the portion of methylated p16 and the portion of p16 protein deficiency by immunohistochemical tests coincided. They stated that p16 could be an important molecular biological marker of cancerous change level of mucosal epithelium and in the diagnosis and progression of oral squamous cell carcinoma. In a similar study, Lopez et al.
22 reported that methylation of the p16 gene was more frequently observed when the level of malignancy was higher at the initial stage of cancerous change.
Similarly, in a study based on the inactivation process of the p16
INK4a gene during the cancerous change of rat tongue cells induced by 4-nitroquinoline 1-oxide, Nakahara et al.
23 observed that the methylation of the p16
INK4a gene was rare in initial and moderate dysplasia but was frequent in severe dysplasia and squamous cell carcinoma. According to Nakahara, inactivation by the hypermethylation of the p16
INK4a gene promoter region is associated with the carcinogenesis of oral cancer.
Gene methylation is observed not only in the p16 gene but also in other genes. Viswanathan et al.
24 studied the methylation of p16, p15, hMLH1, MGMT, and E-cadherin genes associated with oral squamous cell carcinoma and reported that, regardless of the location or pathological stage of the tumor, abnormal methylation did not occur in normal squamous cells but was detected in tumor cells. Viswanathan also stated that the hypermethylation of such genes could be adequately used in the diagnosis of oral squamous cell carcinoma.
In this study, however, the analysis of relations between the immunohistochemical expression of p16 protein and methylation of the p16
INK4a gene and various factors showed that age, gender, smoking habit, and recurrence did not have statistical significance; ditto for the relations between pathological stage, size of tumor, degree of histological differentiation, and survival rate. In particular, methylation was observed in 21 of the 59 tissue samples (35.6%), but there was no correlation with the compared factors. According to Okami et al.
3, there was no statistically signifi-cant relation between methylation level, pathological impression, and prognosis, but the hypermethylation of genes associated with various types of head and neck cancer plays an important role in carcinogenesis. Furthermore, Koscielny et al.
5 reported that the inactivation of p16 in patients with head and neck squamous cell carcinoma was mainly caused by p16 methylation and loss of heterozigosity, whereas cases of point mutation were rare. The correlation between the resulting p16 inactivation and patient survival, metastasis to lymph nodes, and recurrence of tumor was not statistically significant.
In this study, the relation between immunohistochemical p16 expression and methylation of the p16
INK4a gene showed that there is p16 protein expression when p16
INK4a gene methylation occurs; in some cases, p16 protein is not expressed when p16
INK4a gene methylation does not occur. This means that the epigenetic phenomenon of silencing by p16
INK4a gene methylation is not the only cause of suppression of p16 gene expression; other genetic variations such as point mutation and loss of heterozigosity also have an effect. Based on the two-hit theory
25 of Knudson, when methylation occurs on only one side of the paired chromosomes, immunohistochemical results may not show total non-expression even when methylation occurs. Many previous studies reported the correlation between the methylation of the CpG island of patients with oral squamous cell carcinoma and p16 protein expression. According to Huang et al.
26, p16 expression decreases as the methylation of the p16
INK4a gene increases. On the other hand, Ai et al.12 reported that the p16
INK4a gene was hypermethylated in 27% of the genes, but that the expression of p16 protein was suppressed in 74%. In another study, Ai et al.
12 reported that the hypermethylation of the p16
INK4a gene or loss of p16 expression may be referred to as an index of patient survival rate and prognosis evaluation at the initial stage of patients with head and neck squamous cell carcinoma but cannot be used as independent indices of prognosis.
Based on previous studies
2,
3,
5,
12,
24,
26 as well as the results of this study, p16 could be considered methylated in approximately 40% of head and neck squamous cell carcinoma. This implies that p16 methylation cannot be used yet as an index for prognosis evaluation solely but should definitely be considered in the genetic changes of tumor cells along with well-proven factors including the p16/cyclin-dependent kinase/retinoblastoma pathway
12.
The chi-square test results of this study showed signifi-cant correlation between survival rate and histological differentiation level of the tissue (P=0.005). This could be considered an index to verify the reliability of tissue sample extraction and experiment methods applied in this study.
With the development of epigenetics, many studies con-cerning the methylation of genes are in progress. If the role of gene methylation in carcinogenesis could be proven more clearly, it could be applied in various ways including chemotherapy using demethylating drugs
1 and frozen section biopsy of tissue adjacent to the tumor resected during surgery
27.




 PDF
PDF ePub
ePub Citation
Citation Print
Print








 XML Download
XML Download