Abstract
Background
Salvia miltiorrhiza (SM) has been used to treat inflammatory diseases including edema and arthritis; however, the anti-inflammatory mechanism of SM action remains
unresolved.
Methods
The effects of an ethanol extract of SM (ESM) on pro-inflammatory cytokines such as TNF-α, IL-1β, IL-6, and NO, on anti-inflammatory cytokines including IL-4, IL-10, TGF-β, and IL-1Ra have been studied in an attempt to elucidate the anti-inflammatory mechanism in murine macrophages.
Results
ESM inhibited the production of pro-inflammatory cytokines via down-regulation of gene and protein expression whereas it increased the anti-inflammatory cytokines. Furthermore, ESM inhibited the expression of the chemokines, RANTES and CX3CL1, as well as of inflammatory mediators such as TLR-4 and 11β-HSD1.
The immune response is regulated by a highly complex and intricate network of control elements. Cytokines represent the most powerful regulators of immunological reactions. They are direct mediators of inflammation and influence the process and direction of many immunological reactions (1). Overproduction of inflammatory mediators contributes to the pathogenesis of several diseases such as multiple sclerosis, atherosclerosis, rheumatoid arthritis, chronic inflammation, and autoimmune diseases (2-4). Chemokines, a group of cytokines that attracts and activates leukocytes in inflamed tissue, have been associated with the pathogenesis of a number of inflammatory diseases (5).
Some cytokines clearly promote inflammation and are called pro-inflammatory cytokines, whereas others suppress the activity of pro-inflammatory cytokines and thus, are called anti-inflammatory cytokines (1). The pro-inflammatory cytokines, IL-1β, IL-6, and TNF-α mediate and modulate immune response and healing; however, if over-expressed, they may increase the severity of the disease. Anti-inflammatory cytokines, IL-1ra, TGF-β, IL-4, and IL-10 often provide insufficient control over pro-inflammatory activities resulting in immune-mediated diseases. Alternatively, they may overcompensate and inhibit the immune response, leaving the host at risk of systemic infection (6). The functions of Th1 and Th2 cells correlate well with their distinctive cytokines. Th1 cells are involved in cell-mediated inflammatory responses such as the activation of cytotoxic and inflammatory reactions. Alternatively, Th2 cells produce a variety of anti-inflammatory cytokines with negative immunoregulatory effects on cellular immunity that, at the same time, upregulate humoral immunity (7).
Traditionally, pharmacological options for the treatment of inflammatory diseases have been associated with severe side effects (8). Therefore, many scientists continue to look for less toxic efficacious compounds for the treatment of inflammation and cellular injury. Of particular interest is the identification of new anti-inflammatory agents originating from plants used in traditional medicine. For example, Salvia miltiorrhiza (SM) has been commonly used in traditional herbal medicine to prevent and treat vascular diseases such as hypertension, arteriosclerosis, and inflammatory diseases such as edema, arthritis, and endangitis (9-12).
Several studies have shown that SM critically modulates a variety of inflammatory processes and immune functions; however, the molecular mechanisms involved remain to be delineated (13-15). We hypothesize that ESM exerts significant immunomodulatory effects through the modulation of cytokines. The present study was undertaken to investigate the direct effect of ESM on cytokine and chemokine gene and protein expression in murine macrophages.
An extract of 50% ethanol Salvia miltiorrhiza was obtained from Chungbuk National University. Lipopolysaccharide (LPS), and 3-(4,5-dimethylthiazol-2-yl)2,5-dephenyltetrazolium bromide (MTT) were purchased from Sigma (St. Louis, MO, USA). Dulbecco's Modified Eagle Medium (DMEM), antibiotic-penicillin/streptomycin solution, and fetal bovine serum (Hyclone, Logan, UT, USA) were used for cell culture.
RAW 264.7 cells, a murine macrophage cell line, was maintained in a 95% air, 5% CO2 atmosphere in DMEM, supplemented with 10% heat-inactivated fetal bovine serum (FBS), 10,000 U/ml penicillin, and 10,000 U/ml streptomycin.
Cell viability was determined via a MTT reduction assay. In brief, RAW 264.7 cells were pre-incubated overnight in 24-well plates at a density of 2×105 cells per well, and were then washed with 1x PBS. Subsequently, cells were treated with different concentrations of ESM for 24 h and grown in 0.5 mg/ml MTT at 37℃ for 4 h. After removing culture supernatants, the resulting dark blue crystals were dissolved in DMSO (dimethyl sulfoxide). Absorbance values were read at 560 nm using an automated VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA, USA). All determinations were confirmed by replication in at least three independent experiments.
Nitric oxide production from LPS-stimulated RAW 264.7 cells was determined by measuring the amount of nitrite, a relatively stable oxidation product of NO. In brief, RAW 264.7 cells were treated with different concentrations of ESM and then incubated overnight in 96-well plates at a density of 2×105 cells per well. After incubation, 100 µl of culture medium from each sample was mixed with same volume of Griess reagent (stock-I: 0.2% N-(1-naphthyl) ethylenediamine-HCl, stock-II: 2% sulfanilamide in 5% H2PO4). Absorbance values were read at 540 nm. NO concentration was calculated with reference to a standard curve of sodium nitrite, generated using known concentrations.
Assessment of PGE2 synthesis was performed using a commercially available PGE2 Parameter Assay kit (R&D Systems Inc., Minneapolis, MN). RAW 264.7 cells were treated with LPS (100 ng/ml) with different concentrations of ESM for 24 h. PGE2 synthesis was performed on the culture media through a PGE2 enzyme immunoassay using a commercially available kit according to the manufacturer's instruction. The concentration of PGE2 was calculated according to a standard curve using its recombinant protein in the ELISA kit.
The levels of IL-1β, IL-6, and TNF-α in the RAW 264.7 cell culture medium were measured using ELISA kits (eBioscience Inc., San Diego, CA) according to the manufacturer's instructions. The absorbance at 450 nm was determined using a microplate reader (Molecular Devices, Sunnyvale, CA) and the concentration of each cytokine was calculated according to a standard curve using each of the recombinant cytokines in the ELISA kits.
Total RNA was extracted from RAW264.7 cells using the RNeasy Mini kit (QIAGEN, USA) in an RNase-free environment. RNA was quantified by reading the absorbance at 260 nm as previously described (16). The reverse transcription of 1 µg RNA was carried out using M-MLV reverse transcriptase (Promega, USA), oligo (dT) 16 bp primer, dNTP (0.5 µM) and 1 U RNase inhibitor. After incubation at 65℃ for 5 min and 37℃ for 60 min, M-MLV reverse transcriptase was inactivated by heating at 70℃ for 15 min.
The PCR was performed in 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 2.5 mM dNTPs with 5 units of Taq DNA polymerase and 10 pM of each primer set for pro-inflammatory cytokines including IL-1β, IL-6, TNF-α, and cox-2, anti-inflammatory cytokines such as IL-4, IL-10, TGF-β, and IL-1ra, chemokines including CX3CL1, CX3CR1, regulated upon activation normal T-cell expressed and secreted (RANTES), and the inflammatory mediators such as, TLR-4 and 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1). The cDNA was amplified by 35 cycles of denaturing at 94℃ for 45 s, annealing at 60~62℃ for 45 s, and extension at 72℃ for 1 min. Final extension was performed at 72℃ for 5 min.
The PCR products were electrophoresed on 1.5% agarose gels and stained with ethidium bromide (EtBr: 0.5 µg/ml). Verification of specific gene was established by its predicted sizes under ultraviolet (UV) light. The primer sequences were as follows (Forward and Reverse):
iNOS (5'AGCTCCTCCCAGGACCACAC and 5'ACGCTGAGTACCTCATTGGC), COX-2 (5'AAGAAGAAAGTCATTCCTGATCCC and 5'TGACTGTGGGAGGATACATCTCTC), IL-1β (5'CAGGATGAGGACATGACACC and 5'CTCTGCAGACTCAAATCCAC), IL-6 (5'GTACTCCAGAAGACCAGAGG and 5'TGCTGGTGACAACCACGGCC), TNF-α(5'TTGACCTCAGCGCTGAGTTG and 5'CCTGTAGCCCACGTCGTAGC), RANTES (5'ATCATCCTCACTGCAGCCGC and 5'CACACTTGGCGGTTCCTTCG), CX3CL1 (5'CTAAACCAGGACCCTGCGG and 5'CTAGTGGCAAGGTGATCCC), CX3CR1 (5'TCCTGTCCGTCTTCTACGCC and 5'ATTGTGGAGGCCCTCATGGC), TLR4 (5'TGAGAAGTCCCTGCTGAGGC and 5'CTCCTCAGGTCCAAGTTGCC), 11β -HSD1 (5'CAAGGCGGGAAAGCTCATGG and 5' GGAGGAGATGACGGCAATGC), IL-10 (5'TGGCCACACTTGAGAGCTGC and 5'TTCAGGGATGAAGCGGCTGG), TGF-β (5'GGAGCTGGTGAAACGGAAGC and 5'CATAGTAGTCCGCTTCGGGC), and β-actin (5'GTGGGCCGCCCTAGGACCAG and 5'GGAGGAAGAGGATGCGGCAGT). β-actin was used as an internal control.
RAW 264.7 cells were washed with phosphate-buffered saline (PBS) and centrifuged at 1,200 rpm for 10 min at 4℃. Cell pellets were resuspended in an appropriate volume of lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 100 µg/ml phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, and 1% Triton) and incubated overnight at -20℃. After centrifugation at 14,000 rpm for 60 min, the supernatant was stored at -80℃ until further use. Protein concentrations of sample were determined by BCA protein assay (Thermo Fisher Scientific Inc., Rockford, IL). 20 µg/ml of protein from the cell lysates was applied to 8~12% SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk in PBST solution for 1h. They were then incubated with anti-IL-1β, anti-IL-6, anti-TNF-α, anti-i-NOS, anti-COX-2, anti-IL-4, or anti-IL-10 monoclonal antibodies for 2 h and washed 3 times with PBST. After incubation with an alkaline phosphatase-labeled secondary antibody for 2 h, the bands were visualized using a Western Blot Kit with an alkaline phosphatase substrate (Vector, Burlingame, USA).
RAW 264.7 cells (1×106 cells/ml) were cultured in 6-well plates. The cells were treated with various concentrations of ESM and incubated at 37℃ for 24 h in humidified 5% CO2 incubator under standard conditions. The cells then washed with PBS and centrifuged at 1,200 rpm for 10 min at 4℃. The washed cells were blocked with staining buffer containing 10% normal mouse serum (NMS) for 20 min on ice. The blocked cells were then incubated with co-stimulatory molecules such as ICAM-1 (CD 54), B7-1 (CD 80), and B7-2 (CD 86) antibodies for 20 min on ice. Incubated cells were subsequently washed with staining buffer 3 times and then fixed with 1% paraformaldehyde in PBS. The fixed cells were then quantified by flow cytometry (Beckman Coulter, Brea, CA, USA).
Data have been expressed as mean±standard deviation. Statistical significance between the groups was determined by paired t-test and one-way ANOVA for repeated measures. Results with p<0.05 were considered statistically significant. Data were assessed using SPSS (version 15.0, SPSS Inc., Chicago, Illinois).
Initially, the cytotoxicity of ESM to RAW 264.7 cells was measured by MTT assay. The cell viability was not significantly altered by ESM at 12.5~100 µg/ml. Based on these results, cells were treated with ESM in the concentration range of 12.5~100 µg/ml during follow up experiments (data not shown).
In order to determine the effect of ESM on NO production in LPS-stimulated RAW 264.7 cells, the cells were incubated with LPS (100 ng/ml) or various concentrations of ESM (12.5, 25, 50, 100 µg/ml) for 18 h. After incubation, the supernatants were collected and the accumulated nitrite concentration was measured using Griess reagent. Results indicated that macrophages did not release NO in response to the medium alone; LPS (100 ng/ml) was used as a positive control for macrophage activation. Further, ESM was found to inhibit NO production in a dose-dependent manner (Fig. 1A). Also, we determined the effect of ESM on PGE2 production from LPS-stimulated RAW 264.7 cells. LPS alone was able to markedly induce cellular PGE2 production compared to controls. ESM decreased the level of PGE2 production from LPS-stimulated RAW 264.7 cells in a dose-dependent manner (Fig. 1B). ESM also suppressed the mRNA expression and protein induction of iNOS and COX-2 in LPS- stimulated RAW 264.7 cells (Fig. 1C and D). These results suggest that ESM attenuated NO and PGE2 production at the gene and protein levels in activated macrophages.
We then investigated whether ESM inhibited the production of the pro-inflammatory cytokines, IL-1β, IL-6, and TNF-α in LPS-stimulated RAW 264.7 cells. Results indicated that ESM significantly inhibited the production of IL-1β, IL-6, and TNF-α (Fig. 2A). ESM also attenuated their gene expressions and protein induction (Fig. 2B and C). These results suggest that ESM effectively inhibits pro-inflammatory cytokine production through the regulation of gene expression and protein activity of IL-1β, IL-6, and TNF-α in activated macrophages.
Next, we investigated whether ESM inhibited the production of chemokines such as RANTES and CX3CL1 and its receptor, CX3CR1, and inflammatory mediators like TLR-4 and 11β-HSD1. Results demonstrated that ESM significantly decreased the gene expression of RANTES, CX3CL1, CX3CR1, TLR-4, and 11β-HSD1 (Fig. 3), suggesting that ESM effectively inhibits of chemokine production through the regulation of transcription of RANTES, CX3CL1, CX3CR1, TLR-4 and 11β-HSD1 in activated macrophages.
We also investigated whether ESM induces the protein expression of Th2 cytokines such as IL-4, IL-10, and TGF-β. Th2 cytokines are considered anti-inflammatory. ESM enhanced the protein expression of IL-4 and IL-10, and gene induction of IL-10, TGF-β, and IL-1Ra in a dose dependent manner (Fig. 4). These results suggest that ESM increases the productions of anti-inflammatory cytokines through the regulation of transcription of IL-4, IL-10, TGF-β, and IL-1Ra in activated macrophages.
RAW 264.7 cell surface expression of ICAM-1, B7-1, and B7-2 was examined by flow cytometry. As shown in Fig. 5, there were no significant differences in the effect of ESM on ICAM-1, B7-1, or B7-2.
Natural products have played a significant role in drug discovery and development, especially against several well-known diseases. Salvia miltiorrhiza has frequently been used in Chinese medicine as an anti-inflammatory and anti-allergenic agent. A tanshinone derivative was isolated as a major constituent and some of its anti-inflammatory activities were previously described (16-18). Based on this study, we examined the effects of an ethanol extract of Salvia miltiorrhiza (ESM) on pro-inflammatory mediators in LPS- induced murine macrophages. First, we evaluated the cytotoxicity of ESM (12.5~100 µg/ml) in RAW 264.7 cells, the results of which indicated that the tested concentrations of ESM did not affect cell viability (data not shown). Therefore, the inhibition of LPS-induced NO production by ESM was not the result of a cytotoxic effect on these cells.
It has been previously reported that overproduction of inflammatory mediators contributes to the pathogenesis of several diseases such as rheumatoid arthritis, chronic inflammation, and autoimmune diseases. These mediators include numerous chemokines, such RANTES and CX3CL1, and cytokines such TNF-α, IL-1β, and IL-6 (19). Chemokines are a group of chemotactic cytokines that induce cellular locomotion. The rapid production of chemokines at sites of infection leads to the generation of a chemoattractant gradient that enables proper navigation and homing of effector cells (20). RANTES has been reported to attract and activate immune cells and increases in RANTES expression have been associated with a wide range of inflammatory disorders and pathologies, including allogeneic transplant rejection, atherosclerosis, arthritis, and inflammatory airway disorders (21). In all of these pathologies, RANTES has been suggested to act by promoting leukocyte infiltration to sites of inflammation (22). Another chemokine, CX3CL1 is a unique member of the CX3C chemokine subfamily, with each member mediating distinct biological effects (23). Both chemotaxis and adhesion have been shown to be mediated through the G-protein coupled receptor CX3CR1. Accumulating evidence indicates that CX3CL1/CX3CR1 are involved in the pathogenesis of several inflammatory disorders such as glomerulonephritis, rheumatoid arthritis, and systemic lupuserythematosus (24,25). Furthermore, RANTES and CX3CL1 play an important role in the pathogenesis of inflammatory diseases (26-28). In our results, ESM inhibited gene expression of chemokines such as RANTES, CX3CL1, and CX3CR1 (Fig 3A).
Pro-inflammatory cytokines including TNF-α, IL-1β, and IL-6 are present in large quantities in inflammatory tissues. They have been demonstrated to be continuously produced and to be responsible for aggravating inflammation (29). Prostaglandin E2 and NO are important for maintaining inflammatory conditions, and COX-2 and iNOS, which are expressed by inflammatory signals, are the enzymes responsible for producing large amounts of PGE2 and NO. Excessive production of these inflammatory mediators has been detected in septic and hemorrhagic shock, rheumatoid arthritis, and atherosclerosis [4]. According to our results, ESM suppressed the production of NO, PGE2, TNF-α, IL-1β, and IL-6 through a decrease in the gene and protein levels of iNOS, COX-2, TNF-α, IL-1β, and IL-6 in LPS-stimulated RAW 264.7 cells (Figs. 1 and 2).
11β-Hydroxysteroid dehydrogenase type 1 (11β-HSD1) is an intracellular amplifier of glucocorticoids. Glucocorticoids are hormones that regulate a variety of homeostatic processes including metabolism, cell proliferation and differentiation, and immune functions including inflammation (30). Toll-like receptors (TLRs) including TLR1, 2, 4, 5, 6, 7, and 9, play a crucial role in host defense against invading microorganisms. In particular, TLR4 was identified as a signal transducer for LPS, a major cell wall component of Gram-negative bacteria (31). TLR4 signaling led to the induction of pro-inflammatory cytokines through activation of NF-κB. In our results, ESM decreased the expression of inflammatory mediators such as 11β-HSD1 and TLR4 (Fig. 3B).
The most powerful regulatory components are anti-inflammatory cytokines such as IL-4, IL-10, IL-1Ra, and TGF-β. These cytokines either provide insufficient control over pro-inflammatory activities resulting in immune-mediated disease or they overcompensate and inhibit the immune response, rendering the host at risk of systemic infection (1). CD4+ helper cells have been classified as Th1/2-type cells based on the production of specific cytokines (32). The Th2-type cells were reported to produce a variety of anti-inflammatory cytokines, including IL-4, IL-5, and IL-10 (33). These cells derived from IL-4 and IL-10, have been reported to lead to the suppression of Th1 responses by down-regulating the production of macrophages derived from IL-12 and inhibiting the differentiation of Th1-type cells (34). Furthermore, IL-4 has been shown to have marked inhibitory effects on the expression and release to pro-inflammatory cytokines. Additionally, IL-1Ra is known to block the action of IL-1α and IL-1β functional ligands through competitive inhibition at the IL-1 receptor level and TGF-β has been reported to be an important regulator of cell proliferation, differentiation, and formulation of the extracellular matrix. All of these are in clinical trials as anti-inflammatory therapies for inflammatory disease (1). Accordingly, in the current study, the anti-inflammatory effect of ESM in activated RAW 264.7 cells involved the production of anti-inflammatory cytokines including IL-4, IL-10, TGF-β, and IL-1Ra (Fig. 4).
The intracellular adhesion molecules (ICAMs), ICAM-1, ICAM-2 and ICAM-3, are cell surface ligands for the leukocyte integrins. They are crucial in the binding of lymphocytes and other leukocytes to certain cells including antigen-presenting cells (APCs). The B7 family plays important role as co-stimulatory factors of APCs (35). In addition, CD24 combines with the B7 family to aid APCs. Based on our results, ESM did not exhibit significant effects on any of the molecules including ICAM-1, B7- 1, and B7- 2 (Fig. 5). Although the role of co-stimulatory factors is important with regard to the inflammatory reaction, the results here suggest that ESM has anti-inflammatory effects without affecting the expression of ICAMs. In an additional study, ESM did not show significant effects on co-stimulatory molecules with regard to whether different pathways could be regulated by T-cells.
In this study, we have demonstrated that ESM inhibits the production of NO, PGE2, IL-1β, IL-6, and TNF-α, and the expression of RANTES, CX3CL1, CX3CR1, 11β-HSD1, and TLR4, whereas it enhances the production of anti-inflammatory cytokines including IL-4, IL-10, TGF-β, and IL-1Ra in LPS-stimulated macrophages. Thus, we conclude that ESM represents a potentially significant therapeutic drug for the treatment of inflammatory disease.
Figures and Tables
Figure 1
ESM inhibited NO and PGE2 production in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with various concentrations (12.5, 25, 50, 100 µg/ml) in the absence or presence of LPS (100 ng/ml) overnight. Culture supernatants were then collected and NO concentrations were measured using Griess reagent (A). RAW 264.7 cells were treated with various concentrations in the absence or presence of LPS for 48 h. Culture supernatants were then collected and PGE2 concentrations were measured using ELISA kits (B). RAW 264.7 cells were incubated with ESM in the absence or presence of LPS for 24 h. Total RNA was isolated, and levels of iNOS and COX-2 mRNA were then measured by RT-PCR (C). Cell lysates were extracted, and protein levels of COX-2 were then analyzed by Western blotting (D). Each value represents the mean±S.D. in triplicate. ††p<0.01 vs. cells only based on a Student's t-test. *p<0.05, **p<0.01 vs. LPS only based on a student's t-test.
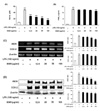
Figure 2
ESM inhibited pro-inflammatory cytokine production in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with various concentrations (12.5, 25, 50, 100µg/ml) in the absence or presence of LPS (100 ng/ml) for 48 h. Culture supernatants were then collected and cytokine concentrations were measured using ELISA kits (A). RAW 264.7 cells were incubated with ESM in the absence or presence of LPS for 24 h. Total RNA was isolated, and mRNA levels of each cytokine were then measured by RT-PCR (B). Cell lysates were extracted, and protein levels of each cytokine were then analyzed by Western blotting (C). Each value represents the mean±S.D. in triplicate. ††p<0.01 vs. cells only based on a Student's t-test. *p<0.05, **p<0.01 vs. LPS only based on a Student's t-test.
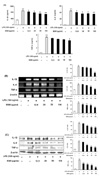
Figure 3
ESM inhibited chemokine production in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with various concentrations (12.5, 25, 50, 100 µg/ml) in the absence or presence of LPS (100 ng/ml) for 24h. Cell lysates were extracted, and protein levels of each chemokine were then analyzed by RT-PCR. Each value represents the mean±S.D. in triplicate. ††p<0.01 vs. cells only based on a Student's t-test. *p<0.05, **p<0.01 vs. LPS only based on a Student's t-test.
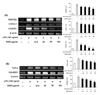
Figure 4
ESM increased Th2 cytokine production in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells were treated with various concentrations (12.5, 25, 50, 100 µg/ml) in the absence or presence of LPS (100 ng/ml) for 24 h. Total RNA was isolated, and mRNA levels of each cytokine were then measured by RT-PCR (A). Cell lysates were extracted, and protein levels of each cytokine were then analyzed by Western blotting (B). Each value represents the mean±S.D. in triplicate. †p<0.05, ††p<0.01 vs. cells only based on a student's t-test. *p<0.05, **p<0.01 vs. LPS only based on a student's t-test.
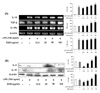
Figure 5
Effects of ESM on the surface expression levels of co-stimulatory molecules. RAW 264.7 cells were cultured and activated with LPS (100 ng/ml) in absence or presence of various ESM concentrations for 24 h. The surface (A) ICAM-1, (B) B7-1, and (C) B7-2 were labeled with either anti-ICAM-1 or anti-B7-1/-2 and the cells were then stained using anti-Vβ8.1+8.2-FITC, anti-Vβ2-PE, or anti-Vβ2-FITC, which served as an isotype control for nonspecific binding.

References
2. Lind L. Circulating markers of inflammation and atherosclerosis. Atherosclerosis. 2003. 169:203–214.

3. Tilg H, Wilmer A, Vogel W, Herold M, Nölchen B, Judmaier G, Huber C. Serum levels of cytokines in chronic liver diseases. Gastroenterology. 1992. 103:264–274.

4. Coker RK, Laurent GJ. Pulmonary fibrosis: cytokines in the balance. Eur Respir J. 1998. 11:1218–1221.

5. Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006. 354:610–621.

7. Nair MP, Kandaswami C, Mahajan S, Chadha KC, Chawda R, Nair H, Kumar N, Nair RE, Schwartz SA. The flavonoid, quercetin, differentially regulates Th-1 (IFNgamma) and Th-2 (IL4) cytokine gene expression by normal peripheral blood mononuclear cells. Biochim Biophys Acta. 2002. 1593:29–36.

8. McKellar G, Madhok R, Singh G. The problem with NSAIDs: what data to believe? Curr Pain Headache Rep. 2007. 11:423–427.

9. Kim JK. Illustrated Natural Drugs Encyclopedia. 1989. Seoul: Namsandan Publishers.
10. Dweck A. Kintzios SE, editor. The folklore and cosmetic use of various Salvia species. Sage The Genus Salvia. 2000. New York: Overseas Publishers;1–25.
11. Duke J, Ayensu E. Medicinal Plants of China. 1985. Algonac, Mich: Reference Publications.
12. Ji XY, Tan BK, Zhu YZ. Salvia miltiorrhiza and ischemic diseases. Acta Pharmacol Sin. 2000. 21:1089–1094.
13. Ding M, Ye TX, Zhao GR, Yuan YJ, Guo ZX. Aqueous extract of Salvia miltiorrhiza attenuates increased endothelial permeability induced by tumor necrosis factor-alpha. Int Immunopharmacol. 2005. 5:1641–1651.

14. Chen TH, Hsu YT, Chen CH, Kao SH, Lee HM. Tanshinone IIA from Salvia miltiorrhiza induces heme oxygenase-1 expression and inhibits lipopolysaccharide-induced nitric oxide expression in RAW 264.7 cells. Mitochondrion. 2007. 7:101–105.

15. Jeon SJ, Son KH, Kim YS, Choi YH, Kim HP. Inhibition of prostaglandin and nitric oxide production in lipopolysaccharide-treated RAW 264.7 cells by tanshinones from the roots of Salvia miltiorrhiza bunge. Arch Pharm Res. 2008. 31:758–763.

16. Kim SY, Moon TC, Chang HW, Son KH, Kang SS, Kim HP. Effects of tanshinone I isolated from Salvia miltiorrhiza bunge on arachidonic acid metabolism and in vivo inflammatory responses. Phytother Res. 2002. 16:616–620.

17. Choi HS, Kim K. Tanshinones inhibit mast cell degranulation by interfering with IgE receptor-mediated tyrosine phosphorylation of PLCg2 and MAPK. Planta Med. 2004. 70:178–180.

18. Jang SI, Jeong SI, Kim KJ, Kim HJ, Yu HH, Park R, Kim HM, You YO. Tanshinone IIA from Salvia miltiorrhiza inhibits inducible nitric oxide synthase expression and production of TNF-, IL-1 and IL-6 in activated RAW 264.7 cells. Planta Med. 2003. 69:1057–1059.

19. Munoz C, Carlet J, Fitting C, Misset B, Blériot JP, Cavaillon JM. Dysregulation of in vitro cytokine production by monocytes during sepsis. J Clin Invest. 1991. 88:1747–1754.

20. Täuber MG, Moser B. Cytokines and chemokines in meningeal inflammation: biology and clinical implications. Clin Infect Dis. 1999. 28:1–11.

21. Hebert CA. Chemokines in disease:biology and clinical research. 1999. Totowa (NJ): Humana Press Inc.
22. Appay V, Rowland-Jones SL. RANTES: a versatile and controversial chemokine. Trends Immunol. 2001. 22:83–87.

23. Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997. 385:640–644.

24. Matsunawa M, Isozaki T, Odai T, Yajima N, Takeuchi HT, Negishi M, Ide H, Adachi M, Kasama T. Increased serum levels of soluble fractalkine (CX3CL1) correlate with disease activity in rheumatoid vasculitis. Arthritis Rheum. 2006. 54:3408–3416.

25. Chen S, Bacon KB, Li L, Garcia GE, Xia Y, Lo D, Thompson DA, Siani MA, Yamamoto T, Harrison JK, Feng L. In vivo inhibition of CC and CX3C chemokine-induced leukocyte infiltration and attenuation of glomerulonephritis in Wistar-Kyoto (WKY) rats by vMIP-II. J Exp Med. 1998. 188:193–198.

26. Schlöndorff D, Nelson PJ, Luckow B, Banas B. Chemokines and renal disease. Kidney Int. 1997. 51:610–621.

27. Schall TJ, Bacon KB. Chemokines, leukocyte trafficking, and inflammation. Curr Opin Immunol. 1994. 6:865–873.

28. Rollins BJ, Yoshimura T, Leonard EJ, Pober JS. Cytokine-activated human endothelial cells synthesize and secrete a monocyte chemoattractant, MCP-1/JE. Am J Pathol. 1990. 136:1229–1233.
29. Choy EH, Panayi GS. Cytokine pathways and joint inflammation in rheumatoid arthritis. N Engl J Med. 2001. 344:907–916.

30. Besedovsky HO, del Rey A. Immune-neuro-endocrine interactions: facts and hypotheses. Endocr Rev. 1996. 17:64–102.

32. Morikawa K, Zhang J, Nonaka M, Morikawa S. Modulatory effect of macrolide antibiotics on the Th1- and Th2-type cytokine production. Int J Antimicrob Agents. 2002. 19:53–59.

33. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986. 136:2348–2357.




 ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download