Dear Editor,
Primary cytogenetic abnormalities of T-cell ALL (T-ALL) frequently involve rearrangements between oncogenic transcription factor genes and T-cell receptor (TCR) loci. These are usually mutually exclusive and associated with specific genetic subgroups [
12]. We report the first case, to our knowledge, of T-ALL showing distinct and rare abnormalities of sequential TCR alpha/delta (
TRA/D) locus rearrangements associated with t(11;14)(p13;q11.2), inv(14)(q11.2q32), and clonal evolution of
JAK2 rearrangement. This report was approved by Asan Medical Center Institutional Review Board, Seoul, Korea (S2019-1375-0001).
A 21-years-old man was admitted to Asan Medical Center in September 2017, with chest discomfort, petechiae, and oral bleeding. The study period was six months from admission. On admission, his hemogram showed a white blood cell count of 55×10
9/L with 68% lymphoblasts, 141 g/L Hb, and 71×10
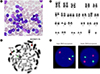
9/L platelets. Computed tomography revealed a mediastinal mass, and a bone marrow (BM) examination showed that 90% of nucleated cells were lymphoblasts (
Fig. 1A). The immunophenotype of the lymphoblasts by four-color flow cytometric analysis (BD FACSCanto II; BD Biosciences, San Jose, CA, USA) was positive for CD2, CD7, CD8, terminal deoxynucleotidyl transferase, and cytoplasmic CD3. The karyotype of the BM cells was 47,XY,del(6)(q13q23),+8,t(11;14)(p13;q11.2),inv(14)(q11.2q32) [13]/46,XY[22] (
Fig. 1B). Metaphase fluorescence
in situ hybridization (FISH) analysis using the Vysis LSI
TRA/D Dual Color Break Apart Rearrangement Probe (Abbott Laboratories, Abbott Park, IL, USA) showed double
TRA/D rearrangements associated with t(11;14) and inv(14) (
Fig. 1C), while interphase FISH revealed a single
TRA/D rearrangement in 12.5% of the cells and double
TRA/D rearrangements in 74.0% of the cells (
Fig. 1D).
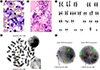
The patient received two cycles of cyclophosphamide, vincristine, doxorubicin, and dexamethasone (hyper-CVAD) for a month and achieved partial remission on day 47. Three weeks later, he was readmitted with disease progression. The follow-up BM examination showed 57.4% lymphoblasts, increased eosinophils (4%), and focal fibrosis (MF-1) (
Fig. 2A and 2B). The BM karyotype was 47,XY,del(6)(q13q23),+8,t(8;9)(p22;p24),t(11;14)(p13; q11.2),inv(14)(q11.2q32)[20]/46,XY[10] including clonal evolution of t(8;9)(p22;p24) (
Fig. 2C). The t(8;9)(p22;p24) component showed a
JAK2 rearrangement by FISH analysis using the JAK2 Break Apart FISH Probe (Empire Genomics LLC, Buffalo, NY, USA) (
Fig. 2D). Metaphase and interphase FISH at diagnosis was negative for
JAK2 rearrangement. Reverse transcriptase-PCR analysis using several primer sets for hotspot breakpoints of
PCM1 (exons 23, 26, 36) and
JAK2 (exons 3, 9, 11, 17) failed to detect the
PCM1-JAK2 fusion transcript; these are most commonly associated with t(8;9)(p22;p24) [
34]. Sequencing of
NOTCH1 revealed an in-frame deletion of the heterodimerization (HD) domain (NM_017617.5:c.4732_4734delGTG (p.V1578del)) and a nonsense mutation in the proline/glutamic acid/serine/threonine (PEST) domain (NM_017617.5:c.5707C>T (p.Q2503*)). We further investigated malignant clones, using a bacterial artificial chromosome FISH probe for RP11-162F6 (Aqua, 11p13), which covers the
LMO2 gene. FISH analysis using the RP11-162F6 and
TRA/D probes showed that
LMO2 was fused with 5′
TRA/D in all cells with a single
TRA/D rearrangement as well as double
TRA/D rearrangements (
Fig. 2E). This indicated that t(11;14) was probably the primary event, rather than inv(14), as a single
TRA/D rearrangement was associated with
LMO2 rearrangement.
The patient was refractory to chemotherapy regimens. He died four months later, after disease progression.
The t(11;14)(p13;q11.2) component is associated with
LMO2 on 11p13 and
TRA/D on 14q11.2 and is found in 6% of T-ALL patients [
1]. Nevertheless, inv(14)(q11.2q32) is very rare in T-ALL. In cases of T-ALL with inv(14)(q11.2q32), possible coexistence with rearrangements of other TCR loci, such as add(7)(q34), t(11;14)(p13;q11.2) and t(8;14)(q24;q11.2), has been reported, indicating that inv(14)(q11.2q32) could constitute a combined cytogenetic abnormality causing T-ALL [
56].
BCL11B and
TRD may be associated with inv(14)(q11.2q32) [
7].
Most cases in a provisional category of myeloid/lymphoid neoplasms associated with t(8;9)(p22;p24.1) and
PCM1-JAK2 are considered myeloid neoplasms, and only a few cases are B-cell ALL (B-ALL) or T-cell lymphoma [
8]. A recent study reported that hematopoietic neoplasms with
JAK2 rearrangement are extremely rare, and most are associated with
PCM1-JAK2 rearrangement [
9]. Our patient, who had a
JAK2 rearrangement of t(8;9)(p22;p24.1) as a clonal evolution, shared the characteristics of the entity with
PCM1-JAK2 rearrangement such as eosinophilia and fibrosis on follow-up BM examination. A
JAK2 inhibitor, such as ruxolitinib, would be effective for treating patients with ALL involving
JAK2 rearrangement [
10].
The t(11;14)(p13;q11.2) component was probably the primary cytogenetic event, with inv(14) and t(8;9) occurring successively. Clonal evolutions, dual NOTCH1 mutations, and additional chromosomal abnormalities may contribute to a rapidly progressive clinical course. Follow-up cytogenetic evaluations and supporting tests, including FISH, are important to detect cytogenetic clonal evolution.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download