Abstract
MATLAB® is widely used for numerical analysis, modeling, and simulation. One of MATLAB's tools, SimBiology®, is often used for pharmacokinetic, pharmacodynamic model and dynamic systems; however, SimBiology seems to be rarely used for non-compartmental analysis (NCA), and the published official documentation provides a poor description of the analysis algorithm for NCA. Therefore, we conducted NCAs with a hypothetical dataset and some scenarios and compared the results. According to the results of this study, SimBiology estimates parameters using the unweighted linear regression for the terminal slope and linear interpolation method. Moreover, although the documentation describing the actual analysis algorithm used to process non-numeric data is not easily accessible to users, users may introduce numeric data at time zero to perform NCA properly. Using the command window, users can perform analyses more quickly and effectively. If the NCA official documentation were improved, SimBiology might be more widely adopted to perform NCA in clinical pharmacology.
Recently, a variety of computer programs have been used for numerical analysis, modeling, and simulation. MATLAB®, which is an abbreviation for ‘matrix laboratory’, is a multi-paradigm numerical computing environment and programming language platform that supports tools for modeling, simulation, visualization, and statistics. Among these tools, SimBiology® provides programmatic tools for modeling, simulation, and analysis of dynamic systems including non-compartmental analysis (NCA), which focuses on pharmacokinetic/pharmacodynamic (PK/PD) and systems biology applications.[1] An NCA is a widely used method for estimating PK/PD parameters, such as the area under the curve (AUC) and clearance without adopting a specific model and it is known to provide more consistent parameter estimates.[2]
SimBiology seems to be rarely used for NCA; a search of PubMed Central® using keywords of ‘SimBiology’ and ‘non-compartmental analysis’ retrieved only seven articles. Contrary to popular belief, in the published user's guide of SimBiology, only 1,055 words address its use for NCA, excluding the description of abbreviations, and also information describing the analysis algorithm is poor.[3] We found a few phrases about the calculation method used for the AUC, “SimBiology uses the linear trapezoidal method to calculate the AUC” and about the regression method of the terminal slope, “to calculate the terminal rate constant (Lambda_z), SimBiology performs a set of linear regressions of the log(concentration)–time data using each of the last n points (n = 3, 4, 5,… ) from the terminal portion of the curve”. Moreover, there is no explicit mention concerning strategies for handling non-numeric data. Therefore, we conducted NCAs with a hypothetical dataset and some scenarios and compared the results.
The hypothetical concentration dataset following an extravascular dose was used to perform NCA with SimBiology (Table 1).[4] Here, terminal slope and AUClast are of interest because other parameters, such as clearance, can be derived from those two parameters and known values. The estimates of terminal slope and AUClast are shown in Table 2. For comparison, we conducted NCAs with several methods for weighting to regression for the terminal slope and for calculation of the AUClast using WinNonlin®, which is widely used for NCA in clinical pharmacology. The results using SimBiology corresponded with the results of uniform weighting and the linear trapezoidal and linear interpolation methods used in WinNonlin.
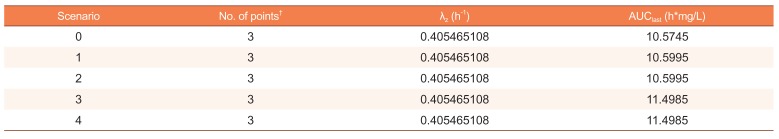
Using SimBiology, we executed various scenarios for handling non-numeric data, such as blanks, characters, or symbols. The scenarios consisted of cases with non-numeric data at time zero or times other than zero (Table 3). The results of those scenarios, including scenario 0 with the original dataset, are shown in Table 4. The AUClast of scenario 1 equaled that of scenario 2 with a linearly interpolated data by points of both sides. This supports that non-numeric data would have been linearly interpolated. Meanwhile, the AUClast of scenario 2 was different from that of the original dataset because the interpolated data was slightly larger than the original numeric data at the same timepoint. The AUClast of scenario 3 (with non-numeric data at time zero) was different from that of the original dataset. The difference was about 8.7%, which may not be acceptable in clinical pharmacology. We also conducted a scenario 4 by replacing non-numeric data at time zero with numeric data at the next closest time point, and the result of scenario 4 was the same as that of scenario 3.
According to the results of this study, SimBiology estimates parameters using the unweighted linear regression for the terminal slope and linear interpolation method. Moreover, although the documentation describing the actual analysis algorithm for handling non-numeric data is not easily accessible to users, users may use datasets with numeric data at time zero to perform NCA properly. SimBiology does have a lower limit of qualification (LLOQ) option that truncates values below the LLOQ to zero; however, that option is neither applicable for non-numeric data nor can it be set separately based on conditions.
We queried the vendor regarding the estimation method used in SimBiology and received the answer that “SimBiology's NCA uses linear regression with all the points weighted equally, that is ‘unweighted’ linear regression.”[5] However, users expect to be able to find explicit descriptions in the published user's guide without having to obtain a personalized answer. Thus, we suggest that the vendor supplement the NCA-related information in their official document by including a detailed explanation for people using or who would like to use SimBiology.
If writing code is a barrier to using SimBiology, a user-friendly interface is available; however, users can perform NCA more quickly, address non-numeric data, and set multiple rules for LLOQ using the command window. Code written in the command window is recorded to a log; thus, it is reproducible and can be tracked. Moreover, because SimBiology runs on MATLAB, the results and the source data can easily be applied to other tools on the MATLAB platform, such as gPKPDsim, which is a graphic user interface application for PK/PD modeling and simulation.[6] Given these advantages, if the NCA-related information in the official documentation were improved, performing NCA with SimBiology might be more widely used in the field of clinical pharmacology.
Acknowledgments
We thank Seokuee Kim, our former colleague, who asked a question and received the answer in the Matlab Answer.
References
1. SimBiology. Accessed 9 September 2019. https://www.mathworks.com/products/simbiology.html.
2. Gabrielsson J, Weiner D. Non-compartmental analysis. Methods Mol Biol. 2012; 929:377–389. DOI: 10.1007/978-1-62703-050-2_16. PMID: 23007438.

3. Noncompartmental Analysis. Accessed 29 August 2019. https://www.mathworks.com/help/simbio/ug/non-compartmental-analysis.html.
4. Rowland M, Tozer TN. Clinical pharmacokinetics/pharmacodynamics. Lippincott Williams and Wilkins Philadelphia;2005.
5. MathWorks-MATLAB Answers. Accessed 5 September 2019. https://kr.mathworks.com/matlabcentral/answers/424217-how-does-simbiology-calculate-the-lambda-in-nca.
6. Hosseini I, Gajjala A, Bumbaca Yadav D, Sukumaran S, Ramanujan S, Paxson R, et al. gPKPDSim: a SimBiology((R))-based GUI application for PKPD modeling in drug development. J Pharmacokinet Pharmacodyn. 2018; 45:259–275. DOI: 10.1007/s10928-017-9562-9. PMID: 29302838.
Table 1
Hypothetical concentration dataset following extravascular dose (modified from ref 4)
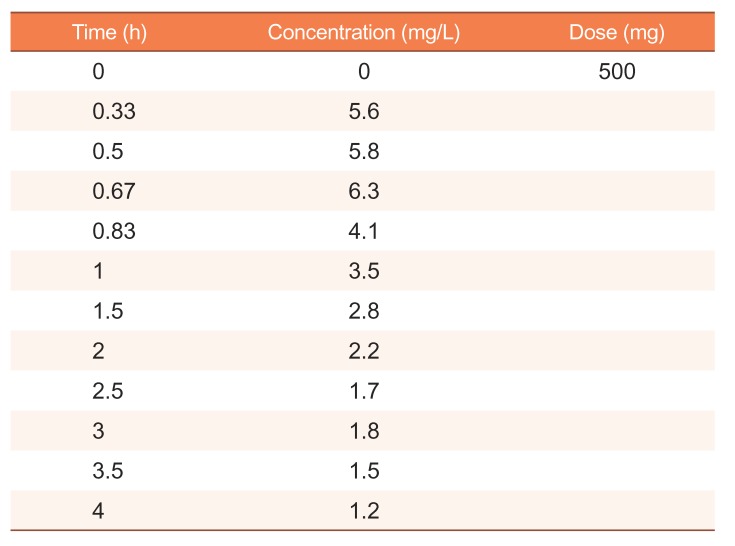
| Time (h) | Concentration (mg/L) | Dose (mg) |
|---|---|---|
| 0 | 0 | 500 |
| 0.33 | 5.6 | |
| 0.5 | 5.8 | |
| 0.67 | 6.3 | |
| 0.83 | 4.1 | |
| 1 | 3.5 | |
| 1.5 | 2.8 | |
| 2 | 2.2 | |
| 2.5 | 1.7 | |
| 3 | 1.8 | |
| 3.5 | 1.5 | |
| 4 | 1.2 |
Table 2
Comparison of non-compartmental analysis results using the hypothetical dataset that does not contain non-numeric data
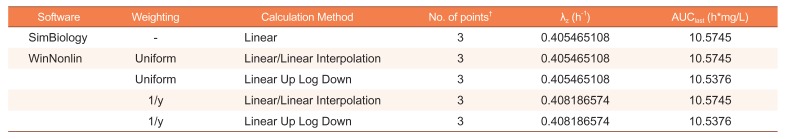
Table 3
List of scenarios for handling non-numeric data





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download