Abstract
Circular RNAs (circRNAs) are currently classed as non-coding RNAs that, unlike the better known canonical linear RNAs, form a covalently closed continuous loop without 5′ or 3′ polarities. With the development of high throughput sequencing technology, a large number of circRNAs have been discovered in many species. More importantly, growing evidence suggests that circRNAs are abundant, evolutionally conserved, and relatively stable in cells and tissues. Strikingly, recent studies have discovered that circRNAs can serve as microRNA sponges, interact with RNA-binding protein, and regulate gene transcription, as well as protein translation. Osteoarthritis (OA) is the most common chronic degenerative joint disease. CircRNAs are differentially expressed in OA cartilage. Moreover, some circRNAs are involved in multiple pathological processes during OA, mainly extracellular matrix degradation, inflammation, and apoptosis. In this review, we briefly delineate the biogenesis, characteristics, and biofunctions of circRNAs, and then, focus on the role of circRNAs in the occurrence and progression OA.
Osteoarthritis (OA) is a chronic degenerative joint disease, primarily characterized by the degradation of articular cartilage.1 OA is more frequent among older adults, commonly affecting peripheral joints, including the knees, hips, and small joints of the hands, and is a leading cause of pain, joint dysfunction, physical disability, substantial morbidity, and reduced quality of life worldwide.2 Multiple factors have been found to be involved in the pathogenesis of OA, including genetic predisposition, altered mechanical loading, and abnormal expression of genes in the articular chondrocytes.3 However, the detailed molecular mechanisms of OA occurrence and progression remain poorly understood, and currently, there are no interventions available to restore degraded cartilage or decelerate disease progression.4 Therefore, it is urgently needed to elucidate the pathological mechanisms of OA and to develop potential alternative therapeutics.
Circular RNAs (circRNAs) are a large class of non-coding RNAs (ncRNAs) that exist ubiquitously in eukaryotic cells;56 however, they have typically been regarded as a byproduct of errant splicing or mRNA process due to low transcript abundance. Only recently, with the rapid development of high throughout RNA sequencing (RNA-Seq) technology and bioinformatics methods, numerous circRNAs have been discovered and identified in human cells, resulting in a resurgence of great interest in the field of genomic research. New evidence suggests that some circRNAs can function as miRNA sponges,789 interact with RNA-binding proteins (RBPs),101112 and regulate gene transcription713 and protein translation.1415 Although studies on circRNAs are still in their infancy, they have emerged as critical players in the occurrence and progression of OA, thereby providing new insights into the underlying molecular mechanisms and treatment of OA.161718 Here, we briefly summarize the classification, biogenesis, characteristics, and biofunctions of circRNAs, and then, review current knowledge on their emerging pathological implications and therapeutic potential in OA.
CircRNAs are mainly divided into three categories: exonic, exon-intron, and intronic circRNA,719 which are produced form different circularizing mechanisms.
Splicing of canonical eukaryotic pre-messenger RNAs (pre-mRNA) is catalyzed by the spliceosomal machinery to remove introns and join exons, leading to formation of a linear RNA transcript with 5′ or 3′ polarity.20 Different from canonical splicing of linear RNA, most circRNAs are generated by a process called backsplicing, which does not follow the canonical 5′-3′ order (Fig. 1).2021 The backsplicing process consists of exon circularization between a downstream 5′ splice site (splice donor) and an upstream 3′ splice site (splice acceptor) in the same pre-mRNA, thereby generating a circular product (circRNAs) without the usual terminal structure {e.g., 5′ cap or a polyadenylated [poly (A)] tail}.72223 As for the mechanism of exon circularization, Jeck and his colleagues7 put forward two models in 2013. One model is termed lariat-driven circularization or exon skipping. A partially folded pre-mRNA transcript brings the original non-adjacent exons close to the others, and then exon skipping occurs, resulting in a crossed region that forms a lariat intermediate containing several exons and introns. Next, the introns in the lariat are removed, generating exonic circRNAs. Generally, introns between the circularized exons are spliced out, although in some cases, they are retained to form exon-intron circRNAs.11 The other model is termed intron-pairing driven circularization or direct backsplicing. Circular structures are formed via base-pairing of ALU complementarity or other RNA secondary structures across flanking introns, resulting in the downstream splice donor being connected to an upstream splice acceptor. Intronic circRNAs are produced from intron lariats that are resistant to degradation by de-branching enzymes.711 Intronic circRNAs contain a single unique 2′-5′ linkage that distinguishes them from exonic circRNAs, and their formation depends on 7 nt GU-rich sequences near the 5′ splice site and 11 nt C-rich sequences close to the branchpoint site.10 Additionally, recent studies have demonstrated another model of circRNAs biogenesis through RBPs. In this case, the alternative splicing factors Quaking protein (QKI)5 and Muscleblind protein (MBL)24 can bind certain circRNAs flanking introns and form a bridge that brings two flanking intronic sequences close together, hence promoting the circularization to form circRNAs. This mechanism is similar to the intron-pairing-driven circularization pathway, except that RBPs regulate adjacent splice sites instead of the direct base pairing between complementary motifs seen in the intron-pairing-driven model.
Along with deep sequencing and bioinformatics, researchers have recognized that circRNAs have several common features. First, circRNAs are abundant. Recent studies have indicated that circRNAs are widely expressed and their expression levels exceed ten times that of the corresponding linear mRNAs in human cells.725 Second, circRNAs are stable. CircRNAs show more stable properties than linear mRNAs in human bodies due to their covalently closed loop structures with neither 5′ cap nor 3′ Poly (A) tails, which confer resistance to RNA exonuclease or ribonuclease R (RNase R) activity.26 This is further substantiated by the average half-life of circRNAs over 48 h, compared to an average half-life of 10 h for mRNAs.27 Third, circRNAs are conserved. Large numbers of circRNAs have highly conserved sequences in different species.725 For example, the circRNA Foxo3 circular RNA (circ-Foxo3) was found to be highly expressed in heart samples from aged patients and mice.28 Fourth, circRNAs are location- and expression-specific. Exonic circRNAs are predominantly in cytoplasm,26 whereas intronic circRNAs are primarily located in the nucleus in eukaryotes, which harbors miRNA binding miRNA response elements (MREs) and forming circRNAs-miRNAs axes are involved in regulation of gene expression at the transcription or post transcription level.710
CircRNAs can function as miRNA sponges, interact with RBPs, and regulate gene transcription, and a few circRNAs can be translated into protein or peptides.
MiRNAs, an abundant class of small ncRNAs (~22 nt), post-transcriptionally regulate the translation of target mRNAs via corresponding MREs.29 Current studies have provided evidence that some circRNAs contain MREs, allowing them to serve as competitive endogenous RNAs to compete for miRNA-binding sites, which leads to the generation of the term “miRNA sponges,” since circRNAs sequester the function of miRNAs so that they can no longer act on their target mRNA.8930 The first miRNA sponge identified was the human circRNA cerebellar degeneration-related protein 1 transcript, which contains 74 binding sites for miR-7, and is, therefore, capable of decreasing miR-7 activity.8 Other circRNAs may function in a similar manner to regulate the activity of particular miRNAs. Murine sex-determining region Y, a testis-specific circRNA, has 16 binding sites for miR-138 and acts as a miR-138 sponge, thereby regulating the expression of miR-138-target genes.9 CircRNA circ-ABCB10 (has-circ-008717) promotes breast cancer proliferation and progression through sponging miR-1271.31 CircHIPK3 serves as a sponge to nine miRNAs with 18 potential binding sites, and directly binds to miR-124, leading to inhibition of miR-124 activity in malignant tumors.32 CirlTCH acts as a miRNA sponge for several miRNAs (miR-7, miR-17, and miR-124) to increase ITCH levels, thereby inhibiting the Wnt/β-catenin pathway.33 In summary, these findings suggest that miRNA sponge effects achieved by circRNA formation may be a general phenomenon,9 and so far, tens of thousands of circRNAs have been found by bioinformatics analysis to exhibit miRNA absorption, which has been found to potentially possess pathological and clinical relevance in human disease.34353637
RBPs may participate in many bioactivities, such as cell proliferation, differentiation, motility, apoptosis, senescence, cellular responses to oxidative stress, and so on. It has been reported that circRNAs can binds multiple RBPs, such as RNA polymerase II (Pol II),10 Argonaute (AGO) proteins,89 MBL,24 QKI,5 eukaryotic initiation factor 4A-III (EIF4A3),38 etc., forming large RNA-protein complexes via stable interaction.39 This interaction with RBPs might lead to a similar effect as miRNA sponging, resulting in depletion of RBPs and a reduction in their interactions with RNA targets.40 For example, MBL can bind exon 2 of its parental gene and promote its circulation to form circMBL, which can then bind MBL to reduce the effective concentrations of MBL and the production of circMBL.24 Recently, a new web tool, CircInteractome (circRNA interactome), has been developed and has helped in the discovery of some circRNAs that are ‘super-sponges’, with an exceptionally high density of binding sites for a given RBP. Among these circRNAs, hsa_circ_0024707 could function as a super sponge for AGO2 with 85 predicted positions, and the mature circRNA hsa_circ_0000020 contains multiple binding sites for several RBPs, such as HuR (6 sites) and FMRP (10 sites).38
Recent studies have shown that some circRNAs can regulate the expression of parental genes.1041 For example, CircEIF3J and circPAIP2, the two most abundant nuclear species of exon-intron circRNAs in HEK-293, contain one potential U1 small nuclear ribonucleoprotein particle (U1snRNP)-binding site in their retained introns, and connect with U1 snRNP to form circRNAs-U1 snRNP complexes that promote interactions with RNA Pol II and the promoter region of the parental gene, resulting in cis transcription initiation of the parental gene.11 Ci-ankrd52, one abundant circRNA derived from the second intron of ANKRD52 gene, accumulates to its sites of transcription, associates with elongation Pol II machinery, and positively regulates Pol II transcription. Knockdown of ciankrd52 leads to the reduced expression of its parental gene.10 From the above, we speculate that circRNAs containing intronic sequences, such as exon-intron circRNAs and intronic circRNA, may regulate transcription in the nucleus, while exon circRNAs may function as miRNA sponges in the cytoplasm.
Because some circRNAs have open reading frames (ORF), it is speculated that they can be translated into proteins. Recently, researchers have uncovered strong evidence for translation of circRNAs, only when circRNAs contain internal ribosome entry site elements (IRES).1542 An IRES is an alternative means of translation initiation in eukaryotes that is independent of the 5′ cap structure and 3′ poly (A) tail recognition. Wang and Wang22 constructed a minigene in vitro, containing the cytomegalovirus promoter, IRES, and an exon encoding green fluorescent protein, which allowed the circularization of corresponding transcription. Legnini, et al.43 identified Circ-ZNF609 with the UTR sequence as an IRES, which is translated into a protein in a splicing-dependent and cap-independent manner. A circRNAs database, named circRNADb, containing 32914 human exonic circRNAs was established to provide detailed information of the circRNAs, including genome sequence, ORF, and IRES, to users for prediction of the translatability of certain circRNAs.14 Yang, et al.44 reported that N6-methyladenosine (m6A) as an IRES promotes efficient initiation of protein translation from circRNAs in human cells. They discovered that m6A motifs are enriched in circRNAs and that a single m6A site is sufficient to drive translation initiation. This m6A-driven translation requires initiation factor eIF4G2 and m6A reader YTHDF3, is enhanced by methyltransferase METTL3/14, inhibited by demethylase FTO, and upregulated upon heat shock. They also found that m6A-driven translation of circRNAs is widespread, with hundreds of endogenous circRNAs having translation potential, suggesting that such translation could be common to many circRNAs. These results challenge the stereotypic view of circRNAs as ncRNAs, and open new paradigms for potential function of circRNAs.
Aberrant expression of circRNAs have been reported to be involved in several types of human diseases, such as cancer, diabetes, cardiovascular disease, and Alzheimer's disease.645 However, the involvement of circRNAs remains largely unknown in the pathogenesis of OA. Recent studies indicated that some circRNAs are aberrantly expressed in human OA cartilage. In a study, Liu, et al.16 discovered that a total of 71 circRNAs were differentially expressed in OA cartilage, compared with normal cartilage. Sixteen of these circRNAs were upregulated and 55 were downregulated in OA tissues. Among these differentially expressed circRNAs, upregulation of circRNA-100876 [chondrocyte extracellular matrix related circRNAs (circRNA-CER)], circRNA_100086, circRNA_101178 and circRNA_101914 was further confirmed by quantitative reverse transcription polymerase chain reaction (qPCR).16 CircRNA-CER was up-regulated in OA with a 2.5-fold change on qPCR. Moreover, the expression of circRNA-CER was upregulated in chondrocyte stimulated with catabolic stimulators, such as interleukin-1 (IL-1) and tumor necrosis factor-α (TNFα).16 Wu, et al.17 found that has-circ-0005105 expression in chondrocytes was significantly upregulated by interleukin-1 beta (IL-1β) in a time dependent manner. Liu, et al.18 identified that a total of 104 circRNAs were differentially expressed in damaged versus intact cartilage. Of these circRNAs, 44 and 60 were upregulated and downregulated, respectively, in the damaged cartilage.18 qRT-PCR results also indicated that circRNA_000598, circRNA_103387, circRNA_101975, and circRNA_100226 [mechanical stress-related circRNAs (circRNAs-MSR)] were overexpressed in the damaged region, compared with the intact region, of cartilage in OA, which is consistent with microarray data.18 Moreover, circRNA-MSR expression increased under mechanical stress in chondrocytes.18 Collectively, there are a number of aberrantly expressed circRNAs in human OA cartilage, and these circRNAs may be involved in the pathogenesis of OA. Further functional investigation of single circRNA is essential to confirm the association with OA and to explore novel potential targets for therapy.
While the underlying molecular mechanisms of OA have not been fully clarified, increased catabolism in the extracellular matrix (ECM) of articular cartilage is acknowledged as a key factor in the occurrence and progression of OA.134 ECM is predominantly composed of type II collagen (ColII) and aggrecan, and the progressive loss of these components is thought to be a primary pathological feature of OA. Matrix-degrading enzymes, such as matrix metalloproteinases (MMPs), and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) are primary enzymes responsible for ECM degradation.3 Moreover, a number of MMPs are highly upregulated in OA cartilage, and knockout of ADAMTS5 leads to significant reduction in the severity of cartilage destruction.3 With the broad biofunctions of circRNAs, it is very possible for some circRNAs to be involved in the regulation of MMPs and ADAMTS expression within articular cartilage.
CircRNA-CER, identified previously as hsa_circ_0023404 in a large effort of RNA-Seq bioinformatics analysis,46 is a special chondrocyte ECM-related circRNA. Its gene is located at chr11:71668272-71671937, and its associated-gene symbol is RNF121. The length of the circRNA-CER is 180 bp. Network analysis of circRNAs-miRNAs-mRNAs interactions was performed, identifying five potential binding sites for miRNA (miR-636, miR-665, miR-217, miR-646, and miR-136) in circRNA-CER.16 Then, the existence of co-regulation of circRNA-CER and MMP13 gene expression upon binding of miR-136 in its 3′UTR region was demonstrated.16 Knockdown of circRNA-CER in OA chondrocytes using small interfering RNA (siRNA) was shown to decrease the expression of mRNA for MMP13 and increase the expression of mRNA for ColII and aggrecan, which were reversed through co-transfection with the miR-136 inhibitor and si-circRNA-CER.16 This occurs mainly because circRNA-CER serves as a miR-136 sponge, sequestering miRNAs and controlling the expression level of MMP13, and participating in the process of chondrocyte ECM degradation.16 Therefore, inhibition of circRNA-CER may provide a novel and promising therapeutic strategy for stimulating ECM regeneration and slowing joint degeneration.
Human has_circ_0005105 contains miR-26a binding site identified by bioinformatics analysis.17 The expression of has_circ_0005105 was significantly upregulated in chondrocytes simulated with IL-1β, whereas the expression of miR-26a in chondrocytes was obviously downregulated, which is negatively correlated with that of has_circ_0005105, indicating that has_circ_0005105 may play a role through miR-26a.17 Quantitative PCR further indicated that has_circ_0005105 could not change the expression of miR-26a, although it could inhibit the transcriptional activity of miR-26a.17 Moreover, Western blot detection showed that has_circ_0005105 could inhibit the expression of ColII and aggrecan, and promote the expression of MMP13 and ADAMTS4, whereas miR-26a has the opposite role, suggesting that has_circ_0005105 can accelerate ECM degradation by regulating the expression of miR-26a.17 In addition, Etich, et al.47 found that dysregulation of miR-26a expression could contribute to ECM changes in cartilage disease and that this miRNA may therefore act as a therapeutic target. Rasheed, et al.48 demonstrated that has-miR-26a-5p may be an important regulator of human cartilage homeostasis and a new target for OA therapy. In a word, these findings suggest that has_circ_0005105 may be an important factor promoting the occurrence and progression of OA.
CircRNA-MSR is a special cartilage circRNA (has_circ_100226, ID circ_0005567 in circBase; http://circbase.org), with its gene located at chr1:51868106-51874004, the symbol of the associated gene being EPS15, and the length of 607 bp. The expression of circRNA-MSR was found to be overexpressed in damaged regions, compared with intact regions, of the cartilage in OA. Moreover, its expression was also significantly increased under mechanical stress in chondrocytes.18 Meanwhile, knockdown of circRNA-MSR could suppress TNF-α expression and increase the expression of ColII and aggrecan,18 indicating that circRNA-MSR can promote ECM degradation. According to the informatics analysis, there are five miRNA-binding sites for circRNA-MSR, and they are miR-138, miR-145, miR-24, miR-620, and miR-875. The circRNA-MSR 3′UTR sequence matchesthese miRNAs, and the TNF-α 3′UTR matches miR-875, suggesting that miR-875 is a common miRNA that can target both circRNA-MSR and TNFα.18 Taken together, circRNA-MSR may promote the degradation of chondrocyte ECM, and knockdown of circRNA-MSR could be a potential therapeutic target for OA.
Has_circ_0045714 was shown to increase the expression of ColII and aggrecan in chondrocytes, while its linear sequences could not.49 Insulin-like growth factor-1 receptor (IGF1R) had similar function; miR-193b could inhibit the expression of ColII and aggrecan.49 IGF1R overexpression could reverse the effect of miR-193b, while miR-193b mimics or IGF1R siRNA could inhibit the function of has_circ_0045714.49 Therefore, has_circ_0045714 may regulate EMC synthesis by promoting the expression of miR-193b target IGF1R.
More and more evidence has shown that inflammation is an important driver of OA cartilage pathology.5051 In addition to traditional proinflammatory mediators, such as IL-1β, TNF-α, and other chemokines, recent studies have implicated nicotinamide phosphoribosyltransferase (NAMPT) in activating OA inflammation pathophysiology.52 NAMPT functions as both an intracellular form (iNAMPT) and an extracellular form (eNAMPT). eNAMPT, also known as visfatin, in human OA chondrocytes inhibits proteoglycan synthesis and increases the expression of matrix-degrading enzymes.53 Recent findings offer a putative role of some circRNAs, in the progression and management of OA inflammation.17 For example, has-circ-0005105 can promote not only the expression NAMPT but also the generation of prostaglandin E2, IL-6, and IL-8, which was due to its regulating the expression of miR-26a.17 Additionally, has-circ-0005105 expression in chondrocytes could be promoted significantly by IL-1β, further suggesting that has-circ-0005105 have some relation to inflammatory factor, IL-1β.17 Liu, et al.18 reported that the silencing of circRNA-MSR by siRNA could suppress TNF-α expression, indicating that circRNA-MSR can promote the expression of TNF-α. In a word, these reports have provided indepth information on a number of inflammation-regulated circRNAs that may be worth pursuing in greater detail with respect to determining their biological functions in regulating chondrocyte inflammation.
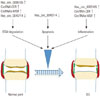
It is well established that decreases in chondrocyte number induced by apoptosis play an important role in the degeneration of cartilage tissues in OA.54 Interestingly, Li, et al.49 reported that has_circ_0045714 could promote chondrocyte proliferation and inhibit chondrocyte apoptosis through the miR-193b target gene IGF1R. Collectively, the above-mentioned results indicated that stimulation of has_circ_0045714 might have a potential therapeutic benefit for OA patients through the blockage of chondrocyte apoptosis. Further investigation, however, is needed to more fully understand the specific mechanisms by which has_circ_0045714 acts to reduce apoptosis. Fig. 2 outlines the roles of circRNAs in the occurrence and development of OA.
RNA interference (RNAi) is a post-transcriptional gene regulation mechanism by which siRNAs induce the sequence-specific degradation of mRNA. RNAi has been achieved in the treatment of cancer,313455 leukemia,56 hepatitis B virus, and human immunodeficiency virus infections.57 Similarly, circRNAs are also able to be silenced by using specific siRNAs. It was reported that the silencing of circRNA-CER or circRNA-MSR by related siRNA can increase chondrocyte ECM formation.1618 Accordingly, circRNA-CER or circRNA-MSR can be used as a potential target, and specific siRNAs can be used as therapeutic agents in OA therapy. The most attractive aspect of siRNAs therapeutics is their high specificity with only one mRNA target, which may make it possible to develop siRNAs-based drugs.58 Thus, circRNAs targeting therapy will surely open up a new therapeutic approach for the treatment of OA. Although this approach is promising, several challenges have been identified in other experiments, including the lack of stability against extracellular and intracellular degradation by nucleases, poor uptake and low potency at target sites of siRNAs, non-specific effects of transfection agents, and off-target effects.5960
With advancements in high throughput sequencing technologies and bioinformatics, investigations of circRNAs are drawing more and more attention. Recent studies suggest that circRNAs serve as new contributors to OA, and also provide novel insights into the pathogenesis of OA. Due to the abundance and stability of circRNAs in vivo, some dysregulated circRNAs may be useful clinical diagnosis biomarkers and therapeutic targets in the future. Recently, a number of circRNAs have been shown to be aberrantly expressed in OA cartilage; however, only four circRNAs have been reported to be involved in OA. Therefore, elucidating the roles of more circRNAs in OA development is urgently needed. Excitedly, circRNA-CER and has_circ_0005105 can function as miRNAs sponges in the pathological state of OA, indicating that some circRNAs may act as upstream regulators of miRNAs. As roles of many miRNAs in the pathogenesis of OA have been verified6162 and several circRNAs have been detected in OA cartilage,16171849 more studies should be done in the future to elucidate the functional connections of circRNAs and miRNAs during OA. With respect to current studies on the effects of circRNAs on OA development, they have all been performed in vitro, and the results thereof need to be validated in vivo. In addition, OA is a chronic degenerative disorder of multifactorial etiology, including chondrocyte ECM degradation,34 chondrocyte apoptosis,63 inflammation,5051 autophagy,6164 oxidative stress,50 and so on. However, recent studies on circRNAs have only been limited to three aspects: ECM degradation, apoptosis, and inflammation in vitro. Therefore, more extensive research has to be done in order to fully understand the biological and molecular mechanisms of circRNAs in the development of OA. Addressing these issues may contribute to disease prevention and the development of therapeutic targets for OA.
Figures and Tables
Fig. 1
Possible biogenesis models of circRNAs. pre-mRNA, pre-messenger RNAs; RBP, RNA-binding protein; circRNAs, circular RNAs.
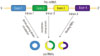
Fig. 2
Roles of circRNAs in the occurrence and development of OA. CircRNAs are regulated in OA cartilage, and associated with ECM degradation, apoptosis, and inflammation of chondrocytes. ↑ indicates upregulation, and ↓ represents downregulation. circRNAs, circular RNAs; OS, osteoarthritis; ECM, extracellular matrix; circRNA-CER, chondrocyte extracellular matrix related circRNAs; circRNAs-MSR, mechanical stress-related circRNAs.
References
1. Rahmati M, Nalesso G, Mobasheri A, Mozafari M. Aging and osteoarthritis: central role of the extracellular matrix. Ageing Res Rev. 2017; 40:20–30.

2. Neogi T. The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage. 2013; 21:1145–1153.

3. Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012; 64:1697–1707.

4. Chen D, Shen J, Zhao W, Wang T, Han L, Hamilton JL, et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 2017; 5:16044.

5. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015; 160:1125–1134.

6. Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017; 4:38.

7. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157.

8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338.

9. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388.

10. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013; 51:792–806.

11. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015; 22:256–264.

12. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015; 18:603–610.

13. Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017; 109:401–407.

14. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016; 6:34985.

15. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016; 1859:1245–1251.

16. Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci Rep. 2016; 6:22572.

17. Wu Y, Zhang Y, Zhang Y, Wang JJ. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol Int. 2017; 41:1283–1289.

18. Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y, et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017; 7:223–230.

19. Hou LD, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017; 30:1–6.

22. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015; 21:172–179.

23. Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015; 6:563–579.

24. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014; 56:55–66.

25. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733.

26. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014; 15:9331–9342.

27. Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342.

28. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017; 38:1402–1412.

29. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014; 15:1–19.

30. Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014; 30:2243–2246.

31. Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017; 7:1566–1576.
32. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016; 7:11215.

33. Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015; 6:6001–6013.

34. Xin Z, Ma Q, Ren S, Wang G, Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics. 2017; 16:80–86.

35. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016; 238:42–51.

36. Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017; 14:992–999.

37. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017; 8:e1386.

38. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016; 13:34–42.

39. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014; 32:453–461.

40. van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci. 2016; 9:74.

41. An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017; 21:185–192.
42. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017; 66:9–21.

43. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017; 66:22–37.

44. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017; 27:626–641.

45. Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of non-coding RNA with novel functions. Exp Biol Med (Maywood). 2017; 242:1136–1141.

46. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013; 9:e1003777.

47. Etich J, Holzer T, Pitzler L, Bluhm B, Brachvogel B. MiR-26a modulates extracellular matrix homeostasis in cartilage. Matrix Biol. 2015; 43:27–34.

48. Rasheed Z, Al-Shobaili HA, Rasheed N, Mahmood A, Khan MI. MicroRNA-26a-5p regulates the expression of inducible nitric oxide synthase via activation of NF-κB pathway in human osteoarthritis chondrocytes. Arch Biochem Biophys. 2016; 594:61–67.

49. Li BF, Zhang Y, Xiao J, Wang F, Li M, Guo XZ, et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 2017; 30:311–318.

50. Marchev AS, Dimitrova PA, Burns AJ, Kostov RV, Dinkova-Kostova AT, Georgiev MI. Oxidative stress and chronic inflammation in osteoarthritis: can NRF2 counteract these partners in crime? Ann N Y Acad Sci. 2017; 1401:114–135.

51. Sun MM, Beier F, Pest MA. Recent developments in emerging therapeutic targets of osteoarthritis. Curr Opin Rheumatol. 2017; 29:96–102.

52. Laiguillon MC, Houard X, Bougault C, Gosset M, Nourissat G, Sautet A, et al. Expression and function of visfatin (Nampt), an adipokine-enzyme involved in inflammatory pathways of osteoarthritis. Arthritis Res Ther. 2014; 16:R38.

53. Yammani RR, Loeser RF. Extracellular nicotinamide phosphoribosyltransferase (NAMPT/visfatin) inhibits insulin-like growth factor-1 signaling and proteoglycan synthesis in human articular chondrocytes. Arthritis Res Ther. 2012; 14:R23.

54. Hwang HS, Kim HA. Chondrocyte apoptosis in the pathogenesis of osteoarthritis. Int J Mol Sci. 2015; 16:26035–26054.

55. Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer. 2017; 16:134.

56. Kolosenko I, Edsbäcker E, Björklund AC, Hamil AS, Goroshchuk O, Grandér D, et al. RNAi prodrugs targeting Plk1 induce specific gene silencing in primary cells from pediatric T-acute lymphoblastic leukemia patients. J Control Release. 2017; 261:199–206.

57. Swamy MN, Wu H, Shankar P. Recent advances in RNAi-based strategies for therapy and prevention of HIV-1/AIDS. Adv Drug Deliv Rev. 2016; 103:174–186.

58. Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015; 4:e252.

59. Barata P, Sood AK, Hong DS. RNA-targeted therapeutics in cancer clinical trials: current status and future directions. Cancer Treat Rev. 2016; 50:35–47.

60. Nabzdyk CS, Pradhan-Nabzdyk L, LoGerfo FW. RNAi therapy to the wall of arteries and veins: anatomical, physiologic, and pharmacological considerations. J Transl Med. 2017; 15:164.

62. D'Adamo S, Cetrullo S, Minguzzi M, Silvestri Y, Borzì RM, Flamigni F. MicroRNAs and autophagy: fine players in the control of chondrocyte homeostatic activities in osteoarthritis. Oxid Med Cell Longev. 2017; 2017:3720128.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download