This article has been
cited by other articles in ScienceCentral.
Abstract
Background
In Korea, there were issues regarding the use of immunoassays for anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies to detect infection. So, we compared antibody results of eight kinds of commercial immunoassays using clinical remnant specimens.
Methods
We compared the results of several immunoassay kits tested on 40 serum samples from 15 confirmed patients and 86 remnant serum samples from clinical laboratory. Eight kinds of IVD kits—four enzyme-linked immunosorbent assay, two lateral flow rapid immunochromatographic assays, and two chemiluminescent immunoassays with one RUO kit were tested.
Results
Among 40 serum samples from 15 coronavirus disease 2019 (COVID-19) patients, 35 yielded at least one positive result for detecting antibodies in the combined assessment. There were inconsistent results in 12 (28%) samples by single immunoassay. Forty samples collected in 2019 before the first COVID-19 Korean case showed negative results except for one equivocal result.
Conclusion
The discrepant results obtained with different immunoassay kits in this study show that serological assessment of SARS-CoV-2 by a single immunoassay requires caution not only in detecting infection but also in assessing immunologic status.
Go to :

Graphical Abstract
Go to :

Keywords: SARS-CoV-2, COVID-19, ELISA, Immunoassay, Korea, Antibodies
INTRODUCTION
Among the countries affected by the global coronavirus disease 2019 (COVID-19) pandemic, the Republic of Korea has controlled the disease relatively well with 506.25 confirmed cases and 8.91 deaths per million, as of October 26, 2020.
12 Only molecular testing has been approved in Korea to diagnose severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. However, in other countries, antibody detection using immunoassays is also accepted for diagnosis. Serological tests for COVID-19 detection have been controversial because of the unique characteristics of immunoassays, diagnostic windows, low sensitivities, and false positives due to cross-reaction or interference.
345 There have been concerns in Korea regarding the use of immunoassays, especially most easy-to-use rapid diagnostic tests, to detect COVID-19 among asymptomatic populations.
6 Using the clinical remnant specimens from the hospitalized patients, we have compared anti-SARS-CoV-2 antibodies results of 8 kinds of available commercial immunoassays, including four assays of two domestic manufacturers, which the U.S. Food and Drug Administration (FDA) had revoked the Emergency Use Authorization.
7
Go to :

METHODS
The clinical information and remnant serum samples of COVID-19 patients confirmed (through respiratory tract samples) by SARS-CoV-2 real-time polymerase chain reaction (PCR) (PowerChek™ 2019-nCoV Real-time PCR Kit; KogeneBiotech, Seoul, Korea) were received by our hospital biobank. For these serum samples, we compared 8 immunoassays including 4 commercial enzyme-linked immunosorbent assay (ELISA) kits. They include spike (S1) protein-based anti-SARS-CoV-2 ELISA (immunoglobulin [Ig] G) and nucleocapsid protein (NCP)-based anti-SARS-CoV-2-NCP ELISA (IgG) (both Euroimmun Ltd., Lübeck, Germany), and 2 Korean manufacturers' kits of standard E COVID-19 total Ab ELISA (SD biosensor, Suwon, Korea) and PCL COVID19 total Ab EIA (PCL, Seoul, Korea) both based on the S protein. Two lateral flow rapid immunochromatographic assays (LFIA), including standard Q COVID-19 IgM/IgG combo test (SD biosensor) targeting the S protein and COVID-19 IgG/IgM rapid gold (PCL) targeting the N protein and the receptor-binding domain of the S protein, and two chemiluminescent immunoassays—VITROS immunodiagnostics products anti-SARS-CoV-2 (S1 and N protein-based) IgG and total (Ortho-Clinical Diagnostics, Inc., Rochester, NY, USA) with complementary assay using cPass™ SARS-CoV-2 neutralization antibody detection RUO kit (GenScript, Inc., Piscataway, NJ, USA).
Forty positive patients' serum samples in were collected from 15 patients who were diagnosed with COVID-19 in our hospital from March 4 to August 7, 2020. Seven male and eight female patients (14 adults aged 29–80 years and a 5-year-old child) were admitted at the collection time of respiratory samples. They were diagnosed within 24 hours as having SARS-CoV-2 infection. Except for 2 asymptomatic patients, a 29-year-old female and a 5-year-old boy, most of them had COVID-19 symptoms, including fever, cough, sputum production, sore throat, myalgia, headache, chills, rhinorrhea, or nasal congestion. Nine patients developed radiologically confirmed pneumonia, and all 15 patients were discharged without mortality. After performing the clinical tests, remnant serum samples were collected from serum separation tubes within 48 hours of drawing blood and frozen at −70°C before the examination.
Negative control samples were also remnant serum samples from the health check-up test in 2019 (n = 40) and the patients (n = 46) with neither SARS-CoV-2 infection nor recent travel history in 2020. The latter serum group was selected from samples with abnormal results in the laboratory tests, which could make cross-reactions in the immunoassay, containing monoclonal paraprotein (n = 4), polyclonal gamma-globulin (n = 2), a high titer of an anti-nuclear antibody (n = 7), or increased serum beta-hCG (n = 3). Patients with positive hepatitis antigen or antibody (n = 5), allergen-specific IgE (n = 1), herpes simplex virus IgM (n = 1), anti-cardiolipin antibody (n = 1), prostate-specific antigen (n = 3), or high level of procalcitonin (n = 7), bilirubin (n = 1), creatinine (n = 2), and serum from the patients positive result for galactomannan (n = 2), reactive particle reagin (n = 4) or Treponema pallidum latex agglutination (n = 1), urinary pneumococcus antigen (n = 1) and respiratory rhinovirus/enterovirus (n = 1) were also included. We tested six assays for the serum samples suspected to be negative, excluding two lateral flow immunoassays, with serum indices measured by VITROS 5600 integrated system (Ortho-Clinical Diagnostics, Inc.). All assays were analyzed according to the manufacturer's instructions and were verified as external quality control materials of other manufacturers' positive (Virotrol SARS-CoV; Bio-Rad Laboratory, Hercules, CA, USA), negative (Viroclear SARS-CoV), and low positive materials (Accurun anti-SARS-CoV-2 reference material kit series 1000; Boston Biomedica, Inc., Cambridge, MA, USA), in addition to the manufacturer's control materials (anti-SARS-CoV-2 total controls and IgG controls; Ortho-Clinical Diagnostics, Inc.).
Ethics statement
This study was reviewed and approved for the deliberation waiver by the Institutional Review Board of Pusan National University Yangsan Hospital (05-2020-017) and was provided with bio-specimens and clinical data from the institutional Biobank Project (OF-2020-10) according to the individual research protocol. Informed consent was waived.
Go to :

RESULTS
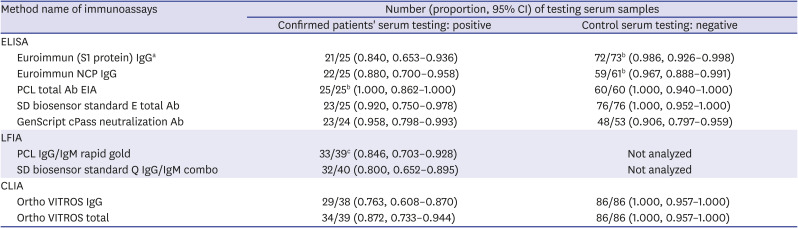
Among 40 serum samples from 15 COVID-19 patients, at least 1 type of anti-SARS-CoV-2 antibody was detected in 35 samples by combining 4 or 8 kinds of immunoassays. In our small group, the clinical sensitivity of each IgG assays showed 76.3%, 84%, and 88% of VITROS IgG, Euroimmun S1, and NCP, respectively (
Table 1). The summed clinical sensitivity of IgG/IgM LFIA was 80% for the SD biosensor and 84.6% for the PCL. These are lower than that of ELISA of same manufacturers (92% for the SD biosensor and 100% for the PCL). 87.2% of the VITROS total antibody by CLIA method was placed between them.
Table 1
Clinical sensitivities and specificities of SARS-CoV-2 antibody detection by immunoassay kits
|
Method name of immunoassays |
Number (proportion, 95% CI) of testing serum samples |
|
Confirmed patients' serum testing: positive |
Control serum testing: negative |
|
ELISA |
|
|
|
Euroimmun (S1 protein) IgGa
|
21/25 (0.840, 0.653–0.936) |
72/73b (0.986, 0.926–0.998) |
|
Euroimmun NCP IgG |
22/25 (0.880, 0.700–0.958) |
59/61b (0.967, 0.888–0.991) |
|
PCL total Ab EIA |
25/25b (1.000, 0.862–1.000) |
60/60 (1.000, 0.940–1.000) |
|
SD biosensor standard E total Ab |
23/25 (0.920, 0.750–0.978) |
76/76 (1.000, 0.952–1.000) |
|
GenScript cPass neutralization Ab |
23/24 (0.958, 0.798–0.993) |
48/53 (0.906, 0.797–0.959) |
|
LFIA |
|
|
|
PCL IgG/IgM rapid gold |
33/39c (0.846, 0.703–0.928) |
Not analyzed |
|
SD biosensor standard Q IgG/IgM combo |
32/40 (0.800, 0.652–0.895) |
Not analyzed |
|
CLIA |
|
|
|
Ortho VITROS IgG |
29/38 (0.763, 0.608–0.870) |
86/86 (1.000, 0.957–1.000) |
|
Ortho VITROS total |
34/39 (0.872, 0.733–0.944) |
86/86 (1.000, 0.957–1.000) |

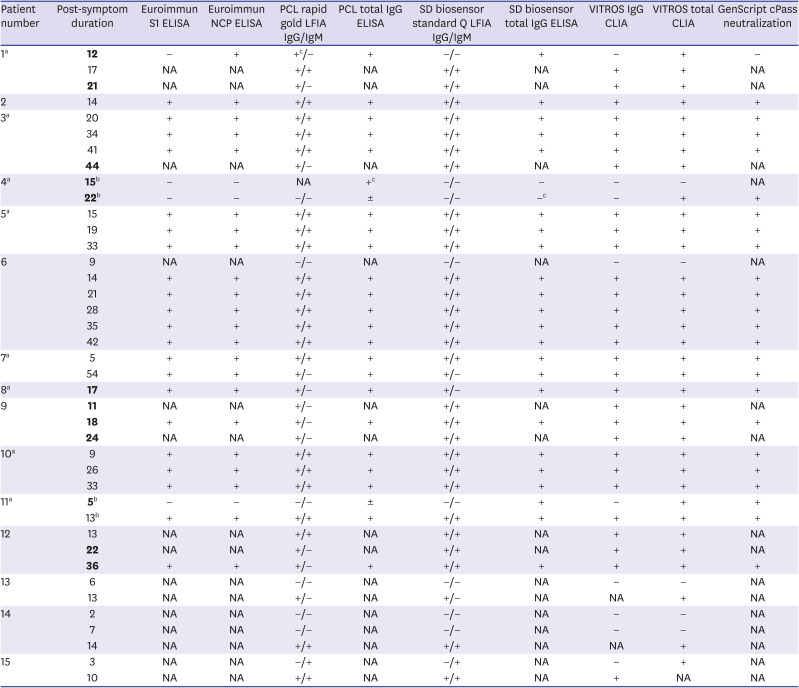
The results were partially inconsistent for 12 (30%) of 40 samples by single assay, including cases where complete evaluation could not be performed because of insufficient reagents. Excluding the most frequent discrepancy—7 results IgM negative in one type of LFIA, 5 samples from 4 patients showed a mismatch between reagents (
Table 2). The reaction signals of 4 assays showed an increasing pattern after symptom onset or infection confirmation in all patients (
Fig. 1). As shown in
Table 2, the comparative results of each sample at different time-points showed very different patterns. In PCL LFIA, IgM results were negative in 7 samples, which was different from the SD biosensor IgM results. The first specimen from patient 1 and two specimens from patient 4 showed three false-suspected results (table footnote c) in a comparison of serial results for the same type of analytes and results of other assays for the same specimen.
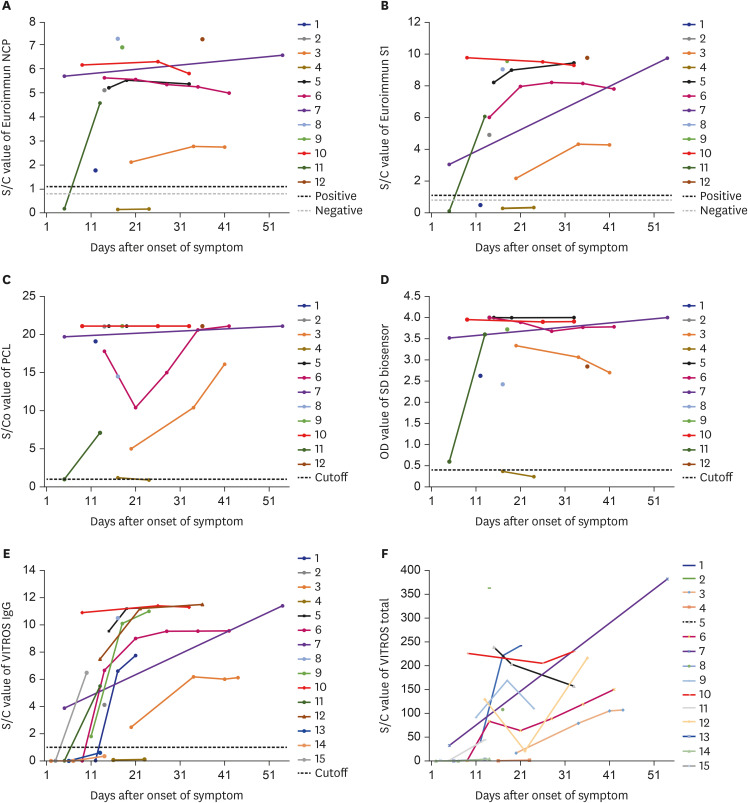 | Fig. 1
Serologic response by reaction values of six immunoassays for anti-SARS-CoV-2 antibody. (A) Euroimmun NCP, (B) S1 IgG EIA, (C) PCL total Ab EIA, (D) SD biosensor standard E total Ab, (E) Ortho VITROS IgG, and (F) Ortho VITROS totala—by duration after symptom onsetb.
SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2, NCP = nucleocapsid protein, S1 = spike, IgG= immunoglobulin G, Ab = antibody, OD = optical density.
aCutoff value (1.0) not displayed due to overlapping of data; bDate after confirmation test for two asymptomatic patients (4 and 11).

|
Table 2
Comparison of SARS-CoV-2 antibody results by immunoassays in serial 40 samples from 15 confirmed COVID-19 patients
|
Patient number |
Post-symptom duration |
Euroimmun S1 ELISA |
Euroimmun NCP ELISA |
PCL rapid gold LFIA IgG/IgM |
PCL total IgG ELISA |
SD biosensor standard Q LFIA IgG/IgM |
SD biosensor total IgG ELISA |
VITROS IgG CLIA |
VITROS total CLIA |
GenScript cPass neutralization |
|
1a
|
12
|
− |
+ |
+c/− |
+ |
−/− |
+ |
− |
+ |
− |
|
17 |
NA |
NA |
+/+ |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
21
|
NA |
NA |
+/− |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
2 |
14 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
3a
|
20 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
34 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
41 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
44
|
NA |
NA |
+/− |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
4a
|
15b
|
− |
− |
NA |
+c
|
−/− |
− |
− |
− |
NA |
|
22b
|
− |
− |
−/− |
± |
−/− |
−c
|
− |
+ |
+ |
|
5a
|
15 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
19 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
33 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
6 |
9 |
NA |
NA |
−/− |
NA |
−/− |
NA |
− |
− |
NA |
|
14 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
21 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
28 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
35 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
42 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
7a
|
5 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
54 |
+ |
+ |
+/− |
+ |
+/− |
+ |
+ |
+ |
+ |
|
8a
|
17
|
+ |
+ |
+/− |
+ |
+/− |
+ |
+ |
+ |
+ |
|
9 |
11
|
NA |
NA |
+/− |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
18
|
+ |
+ |
+/− |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
24
|
NA |
NA |
+/− |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
10a
|
9 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
26 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
33 |
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
11a
|
5b
|
− |
− |
−/− |
± |
−/− |
+ |
− |
+ |
+ |
|
13b
|
+ |
+ |
+/+ |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
12 |
13 |
NA |
NA |
+/+ |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
22
|
NA |
NA |
+/− |
NA |
+/+ |
NA |
+ |
+ |
NA |
|
36
|
+ |
+ |
+/− |
+ |
+/+ |
+ |
+ |
+ |
+ |
|
13 |
6 |
NA |
NA |
−/− |
NA |
−/− |
NA |
− |
− |
NA |
|
13 |
NA |
NA |
+/− |
NA |
+/− |
NA |
NA |
+ |
NA |
|
14 |
2 |
NA |
NA |
−/− |
NA |
−/− |
NA |
− |
− |
NA |
|
7 |
NA |
NA |
−/− |
NA |
−/− |
NA |
− |
− |
NA |
|
14 |
NA |
NA |
+/+ |
NA |
+/+ |
NA |
NA |
+ |
NA |
|
15 |
3 |
NA |
NA |
−/+ |
NA |
−/+ |
NA |
− |
+ |
NA |
|
10 |
NA |
NA |
+/+ |
NA |
+/+ |
NA |
+ |
NA |
NA |

Forty remnant samples collected in 2019 before the first COVID-19 Korean case showed negative results using 6 ELISA or CLIA assays as expected; however, one sample reported an equivocal result in Euroimmun S1 ELISA. Among 46 serum samples from recently hospitalized patients with various disease statuses, which contain high levels of biomarkers that could result in false-positive results, there was only 1 false-positive result in Euroimmun NCP ELISA for serum from a patient diagnosed with plasma cell myeloma. That sample could not be assayed in the VITROS 5600 due to operational error with instrumental flags for the viscous sample and drop error. All assays were analysed according to the manufacturer's instructions and were verified by eight kinds of external quality control materials—two levels (positive and negative materials) for two third-party manufacturers' Virotrol and Viroclear SARS-CoV (Bio-Rad Laboratory) and Accurun anti-SARS-CoV-2 series 1000 (Boston Biomedica, Inc.), and for two kinds from CLIA reagent manufacturer's control materials (anti-SARS-CoV-2 total controls and IgG controls; Ortho-Clinical Diagnostics, Inc.). The coefficient of variation (%) values of positive materials by VITROS IgG and total assays were 2.22%/2.72%for the former two kits, and 3.9%/2.8% for the latter.
Go to :

DISCUSSION
The target product profile has been proposed by the World Health Organization stated that 95%–97% sensitivity and 98%–99% specificity were acceptable and desirable criteria for the diagnosis of COVID-19.
8 This performance was evaluated with automated assays in Public Health England using 536 samples from SARS-CoV-2 infected individuals with ≥ 20 days post-symptom onset. In our study, 13 samples (32.5%) were collected from early-stage COVID-19 patients-six serum samples collected less than 7 days and seven serum samples collected between 7 and 14 days after symptom onset or confirmation by PCR. There were discrepancies in the results of 12 samples from 7 patients using 4–8 different kinds of immunoassays (
Table 2). Only three of them were from the early phase of 14 days, so it is not interpreted as confusion in the early stages of antibody formation. Besides the different antigens targeted by each reagent, many factors can affect the serological assessment of COVID-19. High levels of endogenous components, such as proteins, lipids, or antibodies can interfere with the reaction between the analytes and antigen-specific antibodies in the reagent.
9 There were 9 samples in which the reasons underlying discrepancies could not be determined. In the three false-suspected cases (patient 1 on day 12, patient 4 on day 22, patient 11 on day 5), only VITROS total, CLIA assay which is generally known as more sensitive than ELISA, showed exact positive results that yielded equivocal or non-concordant results using the ELISA total kits. The clinical sensitivities were similar to those reported previously for these assays, but the first antibody detection times varied significantly from patient to patient, 5 to 22 days after symptom onset or infection confirmation.
345101112
Given the small size of the negative control group, clinical specificity was higher than reported in the literature as there was only 1 false-positive case among the 53 to 86 negative control serum samples for each assays. There may be a cutoff issue at a time when reagent upgrades are fast, like these days. GeneScript cPASS neutralization antibody detection RUO kit, used as a complementary assay in this study, showed the lowest clinical specificity due to five false-positive results in the group of serum samples collected before the coronavirus outbreak. Of these, four samples yielded results that were higher than the cut-off value (20% signal inhibition) of the RUO kit and lower than the revised cut-off value of 30% signal inhibition of the FDA-Emergency Use Authorization approved IVD kit.
If the detection of infection would be based on the positive results of a single assay for anti-SARS-CoV-2, there is a risk of misdiagnosis that could cause additional obligation to patients, such as the isolation or molecular diagnostic test for their contacts. The same problem can arise when the majority of the population has been vaccinated against COVID-19, and infections and contention strategies are determined through the detection of antibodies. Additionally, in the clinical setting, other variables like the staff's testing skills and quality management could also affect the test results.
We evaluated patients treated in our hospital located in the middle of the southeast area of Korea and compared with the results from Busan, Ulsan, and Gyeongnam, which showed a very low prevalence of 1.73, 1.39, and 0.925 per 1 million people, respectively, compared to that of 29.35 and 5.34 per 1 million people in the nearby northern area, Daegu and Gyeongsangbuk-do, the largest epidemic area in Korea.
13 If we calculate the clinical performance of immunoassay using the prevalence of COVID-19 (0.07%) by a previous study, even 97% specificity may result in a high false positive rate or very low positive predictive value of 1.9%.
2 To the best of our knowledge, this is the first study in Korea to present antibody results for serial clinical specimens from hospitalized patients, including the results of lateral flow immunoassays. The limitations of this study are the relatively small size, uneven sampling time points, and different combinations of assays. Although some serum samples could not be evaluated for all eight immunoassays due to lack of reagents or samples, the discrepancy in the nearly one-third of the samples shows that the serologic test results for SARS-CoV-2 infection could depend on the reagent selected. We demonstrated that the early antibody pattern of COVID-19 with various commercial assays might help clinicians and laboratorians to select immunoassays. After vaccination commences, clinical sensitivity as well as correlation with neutralizing antibody levels are more important for immune protection. The serological assessment of SARS-CoV-2 infection by a single immunoassay requires caution in the interpretation of positive results and while monitoring the immunologic state.
Go to :

ACKNOWLEDGMENTS
This study was provided with biospecimens and clinical data from the institutional Biobank Project (OF-2020-10) according to the individual research protocol. We would like to thank J Cheon, MT in the Department of Laboratory Medicine, Pusan National University Yangsan Hospital, for the research support.
Go to :

Notes
Go to :

References
1. World Health Organization. WHO coronavirus disease (COVID-19) dashboard. Updated 2020. Accessed October 26, 2020.
https://covid19.who.int.
2. Noh JY, Seo YB, Yoon JG, Seong H, Hyun H, Lee J, et al. Seroprevalence of anti-SARS-CoV-2 antibodies among outpatients in southwestern Seoul, Korea. J Korean Med Sci. 2020; 35(33):e311. PMID:
32830472.

3. Pickering S, Betancor G, Galão RP, Merrick B, Signell AW, Wilson HD, et al. Comparative assessment of multiple COVID-19 serological technologies supports continued evaluation of point-of-care lateral flow assays in hospital and community healthcare settings. PLoS Pathog. 2020; 16(9):e1008817. PMID:
32970782.

4. Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020; 58(7):1081–1088. PMID:
32301749.


5. Favresse J, Eucher C, Elsen M, Laffineur K, Dogné JM, Douxfils J. Response of anti-SARS-CoV-2 total antibodies to nucleocapsid antigen in COVID-19 patients: a longitudinal study. Clin Chem Lab Med. 2020; 58(10):e193–e196. PMID:
32639942.

6. Lee J, Kim SY, Sung H, Choe YJ, Hong KH. Letter to the editor: the interpretation of COVID-19 seroprevalence study should be cautious. J Korean Med Sci. 2020; 35(38):e338. PMID:
32989933.

8. de Lusignan S, Lopez Bernal J, Zambon M, Akinyemi O, Amirthalingam G, Andrews N, et al. Emergence of a novel coronavirus (COVID-19): protocol for extending surveillance used by the Royal College of General Practitioners Research and Surveillance Centre and Public Health England. JMIR Public Health Surveill. 2020; 6(2):e18606. PMID:
32240095.

9. Lifshitz MS. Preanalysis. In : McPherson RA, Pincus MR, editors. Henry's Clinical Diagnosis and Management by Laboratory Methods. 23rd ed. Amsterdam: Elsevier Health Sciences;2017.
10. Choe JY, Kim JW, Kwon HH, Hong HL, Jung CY, Jeon CH, et al. Diagnostic performance of immunochromatography assay for rapid detection of IgM and IgG in coronavirus disease 2019. J Med Virol. 2020; 92(11):2567–2572. PMID:
32458479.


11. Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, et al. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem. 2020; 66(8):1055–1062. PMID:
32402061.


12. Jarrom D, Elston L, Washington J, Prettyjohns M, Cann K, Myles S, et al. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evid Based Med. Forthcoming. 2020; DOI:
10.1136/bmjebm-2020-111511.

13. Ministry of Health and Welfare. Coronavirus disease-19, Republic of Korea. Updated 2020. Accessed October 30, 2020.
http://ncov.mohw.go.kr.
Go to :








 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download